A Numbers Game: Ribosome Densities, Bacterial Growth, and Antibiotic-Mediated Stasis and Death
- PMID: 28174311
- PMCID: PMC5296603
- DOI: 10.1128/mBio.02253-16
A Numbers Game: Ribosome Densities, Bacterial Growth, and Antibiotic-Mediated Stasis and Death
Abstract
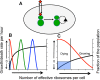
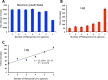
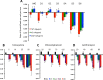
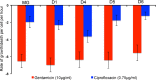
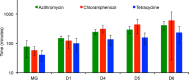
We postulate that the inhibition of growth and low rates of mortality of bacteria exposed to ribosome-binding antibiotics deemed bacteriostatic can be attributed almost uniquely to these drugs reducing the number of ribosomes contributing to protein synthesis, i.e., the number of effective ribosomes. We tested this hypothesis with Escherichia coli K-12 MG1655 and constructs that had been deleted for 1 to 6 of the 7 rRNA (rrn) operons. In the absence of antibiotics, constructs with fewer rrn operons have lower maximum growth rates and longer lag phases than those with more ribosomal operons. In the presence of the ribosome-binding "bacteriostatic" antibiotics tetracycline, chloramphenicol, and azithromycin, E. coli strains with 1 and 2 rrn operons are killed at a substantially higher rate than those with more rrn operons. This increase in the susceptibility of E. coli with fewer rrn operons to killing by ribosome-targeting bacteriostatic antibiotics is not reflected in their greater sensitivity to killing by the bactericidal antibiotic ciprofloxacin, which does not target ribosomes, but also to killing by gentamicin, which does. Finally, when such strains are exposed to these ribosome-targeting bacteriostatic antibiotics, the time before these bacteria start to grow again when the drugs are removed, referred to as the post-antibiotic effect (PAE), is markedly greater for constructs with fewer rrn operons than for those with more rrn operons. We interpret the results of these other experiments reported here as support for the hypothesis that the reduction in the effective number of ribosomes due to binding to these structures provides a sufficient explanation for the action of bacteriostatic antibiotics that target these structures.
Importance: Chemotherapeutic agents, including antibiotics, have been used for more than a century; nevertheless, there are still major gaps in our understanding of how these drugs operate which limit future advances in antibacterial chemotherapy. Although the molecular mechanisms by which antibiotics bind to their target structures are largely known, fundamental questions about how these drugs actually kill and/or inhibit the replication of bacteria remain unanswered and subjects of controversy. We postulate that for the broad class of ribosome-binding bacteriostatic antibiotics, their reducing the number of active (functional) ribosomes per cell provides a sufficient explanation for the abatement of replication and the low rate of decline in densities of viable cells of bacteria exposed to these drugs. Using E. coli K-12 constructs with deletions of from one to six of the seven ribosome-RNA operons and the ribosome-binding bacteriostatic antibiotics tetracycline, chloramphenicol, and azithromycin, we tested this hypothesis. The results of our experiments are consistent with this "numbers game" hypothesis.
Copyright © 2017 Levin et al.
Figures
Similar articles
-
Bactericidal effect of tetracycline in E. coli strain ED1a may be associated with ribosome dysfunction.Nat Commun. 2024 Jun 5;15(1):4783. doi: 10.1038/s41467-024-49084-5. Nat Commun. 2024. PMID: 38839776 Free PMC article.
-
rRNA operon multiplicity as a bacterial genome stability insurance policy.Nucleic Acids Res. 2022 Dec 9;50(22):12601-12620. doi: 10.1093/nar/gkac332. Nucleic Acids Res. 2022. PMID: 35552441 Free PMC article.
-
An examination of the inhibitory effects of three antibiotics in combination on ribosome biosynthesis in Staphylococcus aureus.Arch Microbiol. 2014 Apr;196(4):249-60. doi: 10.1007/s00203-014-0963-5. Epub 2014 Feb 20. Arch Microbiol. 2014. PMID: 24554379
-
Ribosome-targeting antibiotics and mechanisms of bacterial resistance.Nat Rev Microbiol. 2014 Jan;12(1):35-48. doi: 10.1038/nrmicro3155. Nat Rev Microbiol. 2014. PMID: 24336183 Review.
-
Mechanistic Insights into Clinically Relevant Ribosome-Targeting Antibiotics.Biomolecules. 2024 Oct 7;14(10):1263. doi: 10.3390/biom14101263. Biomolecules. 2024. PMID: 39456196 Free PMC article. Review.
Cited by
-
The selection of copiotrophs may complicate biodiversity-ecosystem functioning relationships in microbial dilution-to-extinction experiments.Environ Microbiome. 2023 Mar 17;18(1):19. doi: 10.1186/s40793-023-00478-w. Environ Microbiome. 2023. PMID: 36932455 Free PMC article.
-
Experimental Epidemiology of Antibiotic Resistance: Looking for an Appropriate Animal Model System.Microbiol Spectr. 2018 Feb;6(1):10.1128/microbiolspec.mtbp-0007-2016. doi: 10.1128/microbiolspec.MTBP-0007-2016. Microbiol Spectr. 2018. PMID: 29637886 Free PMC article.
-
Yersinia pseudotuberculosis growth arrest during type-III secretion system expression is associated with altered ribosomal protein expression and decreased gentamicin susceptibility.PLoS Pathog. 2025 Jul 7;21(7):e1012548. doi: 10.1371/journal.ppat.1012548. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40623067 Free PMC article.
-
Macromolecular crowding links ribosomal protein gene dosage to growth rate in Vibrio cholerae.BMC Biol. 2020 Apr 29;18(1):43. doi: 10.1186/s12915-020-00777-5. BMC Biol. 2020. PMID: 32349767 Free PMC article.
-
Interventions on Metabolism: Making Antibiotic-Susceptible Bacteria.mBio. 2017 Nov 28;8(6):e01950-17. doi: 10.1128/mBio.01950-17. mBio. 2017. PMID: 29184022 Free PMC article.
References
-
- Yonath A. 2005. Ribosomal crystallography: peptide bond formation, chaperone assistance and antibiotics activity. Mol Cells 20:1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

