Human-Driven Microbiological Contamination of Benthic and Hyporheic Sediments of an Intermittent Peri-Urban River Assessed from MST and 16S rRNA Genetic Structure Analyses
- PMID: 28174557
- PMCID: PMC5258724
- DOI: 10.3389/fmicb.2017.00019
Human-Driven Microbiological Contamination of Benthic and Hyporheic Sediments of an Intermittent Peri-Urban River Assessed from MST and 16S rRNA Genetic Structure Analyses
Abstract
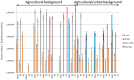
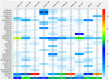
Rivers are often challenged by fecal contaminations. The barrier effect of sediments against fecal bacteria was investigated through the use of a microbial source tracking (MST) toolbox, and by Next Generation Sequencing (NGS) of V5-V6 16S rRNA gene (rrs) sequences. Non-metric multi-dimensional scaling analysis of V5-V6 16S rRNA gene sequences differentiated bacteriomes according to their compartment of origin i.e., surface water against benthic and hyporheic sediments. Classification of these reads showed the most prevalent operating taxonomic units (OTU) to be allocated to Flavobacterium and Aquabacterium. Relative numbers of Gaiella, Haliangium, and Thermoleophilum OTU matched the observed differentiation of bacteriomes according to river compartments. OTU patterns were found impacted by combined sewer overflows (CSO) through an observed increase in diversity from the sewer to the hyporheic sediments. These changes appeared driven by direct transfers of bacterial contaminants from wastewaters but also by organic inputs favoring previously undetectable bacterial groups among sediments. These NGS datasets appeared more sensitive at tracking community changes than MST markers. The human-specific MST marker HF183 was strictly detected among CSO-impacted surface waters and not river bed sediments. The ruminant-specific DNA marker was more broadly distributed but intense bovine pollution was required to detect transfers from surface water to benthic and hyporheic sediments. Some OTU showed distribution patterns in line with these MST datasets such as those allocated to the Aeromonas, Acinetobacter, and Pseudomonas. Fecal indicators (Escherichia coli and total thermotolerant coliforms) were detected all over the river course but their concentrations were not correlated with MST ones. Overall, MST and NGS datasets suggested a poor colonization of river sediments by bovine and sewer bacterial contaminants. No environmental outbreak of these bacterial contaminants was detected.
Keywords: benthic and hyporheic sediments; fecal contamination; high throughput sequencing (HTS); microbial community; peri-urban river.
Figures



Similar articles
-
Ecological assessment of combined sewer overflow management practices through the analysis of benthic and hyporheic sediment bacterial assemblages from an intermittent stream.Sci Total Environ. 2024 Jan 10;907:167854. doi: 10.1016/j.scitotenv.2023.167854. Epub 2023 Oct 15. Sci Total Environ. 2024. PMID: 37848137
-
Characterization and source-tracking of antibiotic resistomes in the sediments of a peri-urban river.Sci Total Environ. 2019 Aug 20;679:88-96. doi: 10.1016/j.scitotenv.2019.05.063. Epub 2019 May 7. Sci Total Environ. 2019. PMID: 31096131
-
Dynamics of toxicity within different compartments of a peri-urban river subject to combined sewer overflow discharges.Sci Total Environ. 2016 Jan 1;539:503-514. doi: 10.1016/j.scitotenv.2015.08.128. Epub 2015 Sep 15. Sci Total Environ. 2016. PMID: 26379263
-
Quantification of Microbial Source Tracking and Pathogenic Bacterial Markers in Water and Sediments of Tiaoxi River (Taihu Watershed).Front Microbiol. 2019 Apr 24;10:699. doi: 10.3389/fmicb.2019.00699. eCollection 2019. Front Microbiol. 2019. PMID: 31105648 Free PMC article.
-
Fecal pollution: new trends and challenges in microbial source tracking using next-generation sequencing.Environ Microbiol. 2018 Sep;20(9):3132-3140. doi: 10.1111/1462-2920.14281. Epub 2018 Aug 5. Environ Microbiol. 2018. PMID: 29797757 Review.
Cited by
-
Ecological assessment of groundwater ecosystems disturbed by recharge systems using organic matter quality, biofilm characteristics, and bacterial diversity.Environ Sci Pollut Res Int. 2020 Jan;27(3):3295-3308. doi: 10.1007/s11356-019-06971-5. Epub 2019 Dec 14. Environ Sci Pollut Res Int. 2020. PMID: 31838704
-
Evidence of Bacterial Community Coalescence between Freshwater and Discharged tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms.Microorganisms. 2023 Apr 2;11(4):922. doi: 10.3390/microorganisms11040922. Microorganisms. 2023. PMID: 37110345 Free PMC article.
-
World's Largest Mass Bathing Event Influences the Bacterial Communities of Godavari, a Holy River of India.Microb Ecol. 2018 Oct;76(3):706-718. doi: 10.1007/s00248-018-1169-1. Epub 2018 Mar 13. Microb Ecol. 2018. PMID: 29536131
-
Bacteriome genetic structures of urban deposits are indicative of their origin and impacted by chemical pollutants.Sci Rep. 2017 Oct 16;7(1):13219. doi: 10.1038/s41598-017-13594-8. Sci Rep. 2017. PMID: 29038457 Free PMC article.
-
Cryptosporidium oocyst persistence in agricultural streams -a mobile-immobile model framework assessment.Sci Rep. 2018 Mar 15;8(1):4603. doi: 10.1038/s41598-018-22784-x. Sci Rep. 2018. PMID: 29545629 Free PMC article.
References
-
- Ashbolt N. J., Grabow W. O., Snozzi M. (2001). Indicators of microbial water quality, in Water Quality: Guidelines, Standards and Health, eds Fewtrell L., Bartram J. (London: World Health Organization; ), 28.
LinkOut - more resources
Full Text Sources
Other Literature Sources

