Comparative Analysis of AGPase Genes and Encoded Proteins in Eight Monocots and Three Dicots with Emphasis on Wheat
- PMID: 28174576
- PMCID: PMC5259687
- DOI: 10.3389/fpls.2017.00019
Comparative Analysis of AGPase Genes and Encoded Proteins in Eight Monocots and Three Dicots with Emphasis on Wheat
Abstract
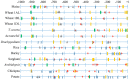
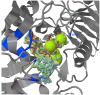
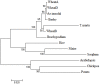
ADP-glucose pyrophosphorylase (AGPase) is a heterotetrameric enzyme with two large subunits (LS) and two small subunits (SS). It plays a critical role in starch biosynthesis. We are reporting here detailed structure, function and evolution of the genes encoding the LS and the SS among monocots and dicots. "True" orthologs of maize Sh2 (AGPase LS) and Bt2 (AGPase SS) were identified in seven other monocots and three dicots; structure of the enzyme at protein level was also studied. Novel findings of the current study include the following: (i) at the DNA level, the genes controlling the SS are more conserved than those controlling the LS; the variation in both is mainly due to intron number, intron length and intron phase distribution; (ii) at protein level, the SS genes are more conserved relative to those for LS; (iii) "QTCL" motif present in SS showed evolutionary differences in AGPase belonging to wheat 7BS, T. urartu, rice and sorghum, while "LGGG" motif in LS was present in all species except T. urartu and chickpea; SS provides thermostability to AGPase, while LS is involved in regulation of AGPase activity; (iv) heterotetrameric structure of AGPase was predicted and analyzed in real time environment through molecular dynamics simulation for all the species; (v) several cis-acting regulatory elements were identified in the AGPase promoters with their possible role in regulating spatial and temporal expression (endosperm and leaf tissue) and also the expression, in response to abiotic stresses; and (vi) expression analysis revealed downregulation of both subunits under conditions of heat and drought stress. The results of the present study have allowed better understanding of structure and evolution of the genes and the encoded proteins and provided clues for exploitation of variability in these genes for engineering thermostable AGPase.
Keywords: ADP_Glucose_PP domain; AGPase; expression analysis; genes structure; ligand binding; molecular dynamics simulation; promoter analysis.
Figures
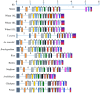
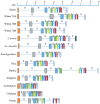

 ), CAAT box (
), CAAT box ( ), light responsive response elements (
), light responsive response elements ( ), abiotic stresses responsive elements (
), abiotic stresses responsive elements ( ), endosperm expression responsive elements (
), endosperm expression responsive elements ( ).
).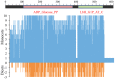
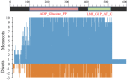


 = transcript (probset id: Ta.2797.2.S1_x_at) encoding AGPase LS and
= transcript (probset id: Ta.2797.2.S1_x_at) encoding AGPase LS and  = transcript (probset id: Ta.242.1.S1_at) encoding AGPase SS. (A) Expression during major abiotic stresses (drought and heat) where fold change is significant at p ≥ 0.05; Numbers on Y-axis indicate 11 different microarray experiments; (B) Expression in different tissues and (C) Expression at different plant developmental stages.
= transcript (probset id: Ta.242.1.S1_at) encoding AGPase SS. (A) Expression during major abiotic stresses (drought and heat) where fold change is significant at p ≥ 0.05; Numbers on Y-axis indicate 11 different microarray experiments; (B) Expression in different tissues and (C) Expression at different plant developmental stages.

References
-
- Bae J. M., Giroux M., Hannah L. C. (1990). Cloning and characterization of the brittle-2 gene of maize. Maydica 35, 317–322.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

