A Comprehensive Analysis of RALF Proteins in Green Plants Suggests There Are Two Distinct Functional Groups
- PMID: 28174582
- PMCID: PMC5258720
- DOI: 10.3389/fpls.2017.00037
A Comprehensive Analysis of RALF Proteins in Green Plants Suggests There Are Two Distinct Functional Groups
Abstract
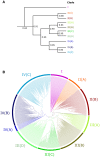
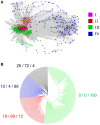
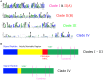
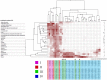
Rapid Alkalinization Factors (RALFs) are small, cysteine-rich peptides known to be involved in various aspects of plant development and growth. Although RALF peptides have been identified within many species, a single wide-ranging phylogenetic analysis of the family across the plant kingdom has not yet been undertaken. Here, we identified RALF proteins from 51 plant species that represent a variety of land plant lineages. The inferred evolutionary history of the 795 identified RALFs suggests that the family has diverged into four major clades. We found that much of the variation across the family exists within the mature peptide region, suggesting clade-specific functional diversification. Clades I, II, and III contain the features that have been identified as important for RALF activity, including the RRXL cleavage site and the YISY motif required for receptor binding. In contrast, members of clades IV that represent a third of the total dataset, is highly diverged and lacks these features that are typical of RALFs. Members of clade IV also exhibit distinct expression patterns and physico-chemical properties. These differences suggest a functional divergence of clades and consequently, we propose that the peptides within clade IV are not true RALFs, but are more accurately described as RALF-related peptides. Expansion of this RALF-related clade in the Brassicaceae is responsible for the large number of RALF genes that have been previously described in Arabidopsis thaliana. Future experimental work will help to establish the nature of the relationship between the true RALFs and the RALF-related peptides, and whether they function in a similar manner.
Keywords: RALF; development; evolution; growth; peptide; phylogeny.
Figures


References
-
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

