Dicer, a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells
- PMID: 28174687
- PMCID: PMC5292673
- DOI: 10.1002/2211-5463.12174
Dicer, a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells
Abstract
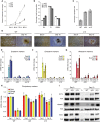
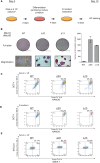
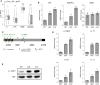
A gene regulation network orchestrates processes ensuring the maintenance of cellular identity and genome integrity. Small RNAs generated by the RNAse III DICER have emerged as central players in this network. Moreover, deletion of Dicer in mice leads to early embryonic lethality. To better understand the underlying mechanisms leading to this phenotype, we generated Dicer-deficient mouse embryonic stem cells (mESCs). Their detailed characterization revealed an impaired differentiation potential, and incapacity to exit from the pluripotency state. We also observed a strong accumulation of LINE-1 (L1s) transcripts, which was translated at protein level and led to an increased L1s retrotransposition. Our findings reveal Dicer as a new essential player that sustains mESCs self-renewal and genome integrity by controlling L1s regulation.
Keywords: Dicer; LINE‐1 retrotransposition; mouse embryonic stem cells; transposable elements.
Figures



References
-
- Bernstein E, Caudy AA, Hammond SM and Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. - PubMed
-
- Foulkes WD, Priest JR and Duchaine TF (2014) DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer 14, 662–672. - PubMed
-
- Svoboda P (2014) Renaissance of mammalian endogenous RNAi. FEBS Lett 588, 2550–2556. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

