The Evolution of LINE-1 in Vertebrates
- PMID: 28175298
- PMCID: PMC5381506
- DOI: 10.1093/gbe/evw247
The Evolution of LINE-1 in Vertebrates
Abstract
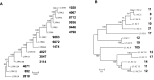
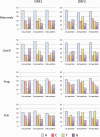
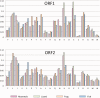
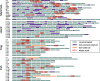
The abundance and diversity of the LINE-1 (L1) retrotransposon differ greatly among vertebrates. Mammalian genomes contain hundreds of thousands L1s that have accumulated since the origin of mammals. A single group of very similar elements is active at a time in mammals, thus a single lineage of active families has evolved in this group. In contrast, non-mammalian genomes (fish, amphibians, reptiles) harbor a large diversity of concurrently transposing families, which are all represented by very small number of recently inserted copies. Why the pattern of diversity and abundance of L1 is so different among vertebrates remains unknown. To address this issue, we performed a detailed analysis of the evolution of active L1 in 14 mammals and in 3 non-mammalian vertebrate model species. We examined the evolution of base composition and codon bias, the general structure, and the evolution of the different domains of L1 (5′UTR, ORF1, ORF2, 3′UTR). L1s differ substantially in length, base composition, and structure among vertebrates. The most variation is found in the 5′UTR, which is longer in amniotes, and in the ORF1, which tend to evolve faster in mammals. The highly divergent L1 families of lizard, frog, and fish share species-specific features suggesting that they are subjected to the same functional constraints imposed by their host. The relative conservation of the 5′UTR and ORF1 in non-mammalian vertebrates suggests that the repression of transposition by the host does not act in a sequence-specific manner and did not result in an arms race, as is observed in mammals.
Figures






References
-
- Adey NB, Schichman SA, et al. 1994. Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol Biol Evol. 11:778–789. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

