Overexpression of antibiotic resistance genes in hospital effluents over time
- PMID: 28175320
- PMCID: PMC5437528
- DOI: 10.1093/jac/dkx017
Overexpression of antibiotic resistance genes in hospital effluents over time
Abstract
Objectives: Effluents contain a diverse abundance of antibiotic resistance genes that augment the resistome of receiving aquatic environments. However, uncertainty remains regarding their temporal persistence, transcription and response to anthropogenic factors, such as antibiotic usage. We present a spatiotemporal study within a river catchment (River Cam, UK) that aims to determine the contribution of antibiotic resistance gene-containing effluents originating from sites of varying antibiotic usage to the receiving environment.
Methods: Gene abundance in effluents (municipal hospital and dairy farm) was compared against background samples of the receiving aquatic environment (i.e. the catchment source) to determine the resistome contribution of effluents. We used metagenomics and metatranscriptomics to correlate DNA and RNA abundance and identified differentially regulated gene transcripts.
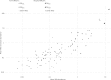
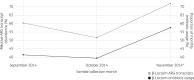
Results: We found that mean antibiotic resistance gene and transcript abundances were correlated for both hospital ( ρ = 0.9, two-tailed P <0.0001) and farm ( ρ = 0.5, two-tailed P <0.0001) effluents and that two β-lactam resistance genes ( bla GES and bla OXA ) were overexpressed in all hospital effluent samples. High β-lactam resistance gene transcript abundance was related to hospital antibiotic usage over time and hospital effluents contained antibiotic residues.
Conclusions: We conclude that effluents contribute high levels of antibiotic resistance genes to the aquatic environment; these genes are expressed at significant levels and are possibly related to the level of antibiotic usage at the effluent source.
© The Author 2017. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures

References
-
- Holmes AH, Moore LSP, Sundsfjord A. et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016; 387: 176–87. - PubMed
-
- D’Costa VM, McGrann KM, Hughes DW. et al. Sampling the antibiotic resistome. Science 2006; 311: 374–7. - PubMed
-
- Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 2009; 157: 2893–902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

