Nanodiscs in Membrane Biochemistry and Biophysics
- PMID: 28177242
- PMCID: PMC5805400
- DOI: 10.1021/acs.chemrev.6b00690
Nanodiscs in Membrane Biochemistry and Biophysics
Abstract
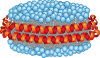
Membrane proteins play a most important part in metabolism, signaling, cell motility, transport, development, and many other biochemical and biophysical processes which constitute fundamentals of life on the molecular level. Detailed understanding of these processes is necessary for the progress of life sciences and biomedical applications. Nanodiscs provide a new and powerful tool for a broad spectrum of biochemical and biophysical studies of membrane proteins and are commonly acknowledged as an optimal membrane mimetic system that provides control over size, composition, and specific functional modifications on the nanometer scale. In this review we attempted to combine a comprehensive list of various applications of nanodisc technology with systematic analysis of the most attractive features of this system and advantages provided by nanodiscs for structural and mechanistic studies of membrane proteins.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

