Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function
- PMID: 28177435
- PMCID: PMC6246501
- DOI: 10.1093/annonc/mdw437
Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function
Abstract
Background: Lung squamous cell carcinoma (LUSC) accounts for 20–30% of non-small cell lung cancers (NSCLCs). There are limited treatment strategies for LUSC in part due to our inadequate understanding of the molecular underpinnings of the disease. We performed whole-exome sequencing (WES) and comprehensive immune profiling of a unique set of clinically annotated early-stage LUSCs to increase our understanding of the pathobiology of this malignancy.
Methods: Matched pairs of surgically resected stage I-III LUSCs and normal lung tissues (n = 108) were analyzed by WES. Immunohistochemistry and image analysis-based profiling of 10 immune markers were done on a subset of LUSCs (n = 91). Associations among mutations, immune markers and clinicopathological variables were statistically examined using analysis of variance and Fisher’s exact test. Cox proportional hazards regression models were used for statistical analysis of clinical outcome.
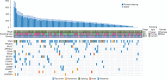
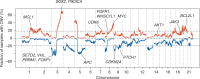
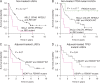
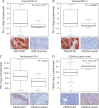
Results: This early-stage LUSC cohort displayed an average of 209 exonic mutations per tumor. Fourteen genes exhibited significant enrichment for somatic mutation: TP53, MLL2, PIK3CA, NFE2L2, CDH8, KEAP1, PTEN, ADCY8, PTPRT, CALCR, GRM8, FBXW7, RB1 and CDKN2A. Among mutated genes associated with poor recurrence-free survival, MLL2 mutations predicted poor prognosis in both TP53 mutant and wild-type LUSCs. We also found that in treated patients, FBXW7 and KEAP1 mutations were associated with poor response to adjuvant therapy, particularly in TP53-mutant tumors. Analysis of mutations with immune markers revealed that ADCY8 and PIK3CA mutations were associated with markedly decreased tumoral PD-L1 expression, LUSCs with PIK3CA mutations exhibited elevated CD45ro levels and CDKN2A-mutant tumors displayed an up-regulated immune response.
Conclusion(s): Our findings pinpoint mutated genes that may impact clinical outcome as well as personalized strategies for targeted immunotherapies in early-stage LUSC.
Figures
Comment in
-
'Genotype/immunotype' correlations in resected NSCLC.Ann Oncol. 2017 Jan 1;28(1):7-8. doi: 10.1093/annonc/mdw624. Ann Oncol. 2017. PMID: 28039156 Free PMC article. No abstract available.
-
Genomics of lung cancer.J Thorac Dis. 2017 Feb;9(2):E155-E157. doi: 10.21037/jtd.2017.02.29. J Thorac Dis. 2017. PMID: 28275503 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

