Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis
- PMID: 28177907
- PMCID: PMC5410358
- DOI: 10.18632/oncotarget.15074
Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis
Abstract
Background: The number of studies on the association between clock genes' polymorphisms and cancer susceptibility has increased over the last years but the results are often conflicting and no comprehensive overview and quantitative summary of the evidence in this field is available.
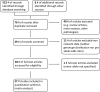
Results: Literature search identified 27 eligible studies comprising 96756 subjects (cases: 38231) and investigating 687 polymorphisms involving 14 clock genes. Overall, 1025 primary and subgroup meta-analyses on 366 gene variants were performed. Study distribution by tumor was as follows: breast cancer (n=15), prostate cancer (n=3), pancreatic cancer (n=2), non-Hodgkin's lymphoma (n=2), glioma (n=1), chronic lymphocytic leukemia (n=1), colorectal cancer (n=1), non-small cell lung cancer (n=1) and ovarian cancer (n=1).We identified 10 single nucleotide polymorphisms (SNPs) significantly associated with cancer risk: NPAS2 rs10165970 (mixed and breast cancer shiftworkers), rs895520 (mixed), rs17024869 (breast) and rs7581886 (breast); CLOCK rs3749474 (breast) and rs11943456 (breast); RORA rs7164773 (breast and breast cancer postmenopausal), rs10519097 (breast); RORB rs7867494 (breast cancer postmenopausal), PER3 rs1012477 (breast cancer subgroups) and assessed the level of quality evidence to be intermediate. We also identified polymorphisms with lower quality statistically significant associations (n=30).
Conclusions: Our work supports the hypothesis that genetic variation of clock genes might affect cancer risk. These findings also highlight the need for more efforts in this research field in order to fully establish the contribution of clock gene variants to the risk of developing cancer.
Methods: We conducted a systematic review and meta-analysis of the evidence on the association between clock genes' germline variants and the risk of developing cancer. To assess result credibility, summary evidence was graded according to the Venice criteria and false positive report probability (FPRP) was calculated to further validate result noteworthiness. Subgroup meta-analysis was also performed based on participant features and tumor type. The breast cancer subgroup was further stratified by work conditions, estrogen receptor/progesterone receptor status and menopausal status, conditions associated with the risk of breast cancer in different studies.
Keywords: SNP; cancer risk; circadian rhythms; clock genes; meta-analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Foster RG, Kreitzman L. The rhythms of life: what your body clock means to you! Experimental physiology. 2014;99:599–606. - PubMed
-
- Merbitz-Zahradnik T, Wolf E. How is the inner circadian clock controlled by interactive clock proteins?: Structural analysis of clock proteins elucidates their physiological role. FEBS letters. 2015;589:1516–1529. - PubMed
-
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. - PubMed
-
- Ercolani L, Ferrari A, De Mei C, Parodi C, Wade M, Grimaldi B. Circadian clock: Time for novel anticancer strategies? Pharmacological research. 2015;100:288–295. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

