Metabolomic signatures associated with disease severity in multiple sclerosis
- PMID: 28180139
- PMCID: PMC5278923
- DOI: 10.1212/NXI.0000000000000321
Metabolomic signatures associated with disease severity in multiple sclerosis
Abstract
Objective: To identify differences in the metabolomic profile in the serum of patients with multiple sclerosis (MS) compared to controls and to identify biomarkers of disease severity.
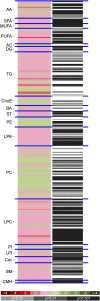
Methods: We studied 2 cohorts of patients with MS: a retrospective longitudinal cohort of 238 patients and 74 controls and a prospective cohort of 61 patients and 41 controls with serial serum samples. Patients were stratified into active or stable disease based on 2 years of prospective assessment accounting for presence of clinical relapses or changes in disability measured with the Expanded Disability Status Scale (EDSS). Metabolomic profiling (lipids and amino acids) was performed by ultra-high-performance liquid chromatography coupled to mass spectrometry in serum samples. Data analysis was performed using parametric methods, principal component analysis, and partial least square discriminant analysis for assessing the differences between cases and controls and for subgroups based on disease severity.
Results: We identified metabolomics signatures with high accuracy for classifying patients vs controls as well as for classifying patients with medium to high disability (EDSS >3.0). Among them, sphingomyelin and lysophosphatidylethanolamine were the metabolites that showed a more robust pattern in the time series analysis for discriminating between patients and controls. Moreover, levels of hydrocortisone, glutamic acid, tryptophan, eicosapentaenoic acid, 13S-hydroxyoctadecadienoic acid, lysophosphatidylcholines, and lysophosphatidylethanolamines were associated with more severe disease (non-relapse-free or increase in EDSS).
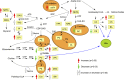
Conclusions: We identified metabolomic signatures composed of hormones, lipids, and amino acids associated with MS and with a more severe course.
Figures

References
-
- Ibrahim SM, Gold R. Genomics, proteomics, metabolomics: what is in a word for multiple sclerosis? Curr Opin Neurol 2005;18:231–235. - PubMed
-
- Villoslada P. Biomarkers for multiple sclerosis. Drug News Perspect 2010;23:585–595. - PubMed
-
- Villoslada P, Baranzini S. Data integration and systems biology approaches for biomarker discovery: challenges and opportunities for multiple sclerosis. J Neuroimmunol 2012;248:58–65. - PubMed
-
- Comabella M, Montalban X. Body fluid biomarkers in multiple sclerosis. Lancet Neurol 2014;13:113–126. - PubMed
-
- Tumani H, Hartung HP, Hemmer B, et al. . Cerebrospinal fluid biomarkers in multiple sclerosis. Neurobiol Dis 2009;35:117–127. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
