Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH
- PMID: 28180276
- PMCID: PMC5605235
- DOI: 10.1093/nar/gkx090
Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH
Erratum in
-
Corrigendum: Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH.Nucleic Acids Res. 2017 Dec 15;45(22):13095-13096. doi: 10.1093/nar/gkx1178. Nucleic Acids Res. 2017. PMID: 29140476 Free PMC article. No abstract available.
Abstract
i-Motifs are alternative DNA secondary structures formed in cytosine-rich sequences. Particular examples of these structures, traditionally assumed to be stable only at acidic pH, have been found to form under near-physiological conditions. To determine the potential impact of these structures on physiological processes, investigation of sequences with the capacity to fold under physiological conditions is required. Here we describe a systematic study of cytosine-rich DNA sequences, with varying numbers of consecutive cytosines, to gain insights into i-motif DNA sequence and structure stability. i-Motif formation was assessed using ultraviolet spectroscopy, circular dichroism and native gel electrophoresis. We found that increasing cytosine tract lengths resulted in increased thermal stability; sequences with at least five cytosines per tract folded into i-motif at room temperature and neutral pH. Using these results, we postulated a folding rule for i-motif formation, analogous to (but different from) that for G-quadruplexes. This indicated that thousands of cytosine-rich sequences in the human genome may fold into i-motif structures under physiological conditions. Many of these were found in locations where structure formation is likely to influence gene expression. Characterization of a selection of these identified i-motif forming sequences uncovered 17 genomic i-motif forming sequence examples which were stable at neutral pH.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
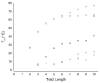
 pH 7.4 Tm1; × pH 7.4 Tm2; and ■ pH 7.4 Ta.
pH 7.4 Tm1; × pH 7.4 Tm2; and ■ pH 7.4 Ta.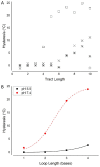
 Secondary hysteresis at pH 7.4. (B) Hysteresis at pH 5.5 (▪) and at pH 7.4 (•) in ODNs containing tracts of 5 cytosines with increasing lengths of thymine loops.
Secondary hysteresis at pH 7.4. (B) Hysteresis at pH 5.5 (▪) and at pH 7.4 (•) in ODNs containing tracts of 5 cytosines with increasing lengths of thymine loops.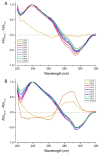
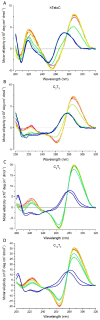
 pH 4.0;
pH 4.0;  pH 4.5;
pH 4.5;  pH 5.0;
pH 5.0;  pH 5.5;
pH 5.5;  pH 6.0;
pH 6.0;  pH 6.5;
pH 6.5;  pH 7.0;
pH 7.0;  pH 7.5;
pH 7.5;  pH 8.0. All ODN concentrations were 10 μM in 10 mM sodium cacodylate buffer with 100 mM sodium chloride buffer at the required pH.
pH 8.0. All ODN concentrations were 10 μM in 10 mM sodium cacodylate buffer with 100 mM sodium chloride buffer at the required pH.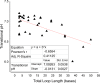

References
-
- Huppert J.L. Hunting G-quadruplexes. Biochimie. 2008; 90:1140–1148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

