Dynamic chromatin accessibility modeled by Markov process of randomly-moving molecules in the 3D genome
- PMID: 28180283
- PMCID: PMC5449544
- DOI: 10.1093/nar/gkx086
Dynamic chromatin accessibility modeled by Markov process of randomly-moving molecules in the 3D genome
Abstract
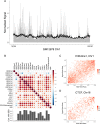
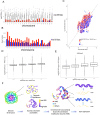
Chromatin three-dimensional (3D) structure plays critical roles in gene expression regulation by influencing locus interactions and accessibility of chromatin regions. Here we propose a Markov process model to derive a chromosomal equilibrium distribution of randomly-moving molecules as a functional consequence of spatially organized genome 3D structures. The model calculates steady-state distributions (SSD) from Hi-C data as quantitative measures of each chromatin region's dynamic accessibility for transcription factors and histone modification enzymes. Different from other Hi-C derived features such as compartment A/B and interaction hubs, or traditional methods measuring chromatin accessibility such as DNase-seq and FAIRE-seq, SSD considers both chromatin-chromatin and protein-chromatin interactions. Through our model, we find that SSD could capture the chromosomal equilibrium distributions of activation histone modifications and transcription factors. Compared with compartment A/B, SSD has higher correlations with the binding of these histone modifications and transcription factors. In addition, we find that genes located in high SSD regions tend to be expressed at higher level. Furthermore, we track the change of genome organization during stem cell differentiation, and propose a two-stage model to explain the dynamic change of SSD and gene expression during differentiation, where chromatin organization genes first gain chromatin accessibility and are expressed before lineage-specific genes do. We conclude that SSD is a novel and better measure of dynamic chromatin activity and accessibility.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lanctôt C., Cheutin T., Cremer M., Cavalli G., Cremer T.. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007; 8:104–115. - PubMed
-
- Cohen B.A., Mitra R.D., Hughes J.D., Church G.M.. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 2000; 26:183–186. - PubMed
-
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C. et al. . Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006; 16:1299–1309. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

