A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron
- PMID: 28180295
- PMCID: PMC5388394
- DOI: 10.1093/nar/gkw1051
A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron
Abstract
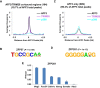
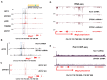
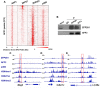
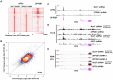
Genomic imprinting is an epigenetic regulation that leads to gene expression in a parent-of-origin specific manner. AFF3, the central component of the Super Elongation Complex-like 3 (SEC-L3), is enriched at both the intergenic-differentially methylated region (IG-DMR) and the Meg3 enhancer within the imprinted Dlk1-Dio3 locus to regulate the allele-specific gene expression in this locus. The localization of AFF3 to IG-DMR requires ZFP57. However, how AFF3 functions at the Meg3 enhancer in maintaining allele-specific gene expression remains unclear. Here, we demonstrate that AFF3 is associated with the Krüppel-like zinc finger protein ZFP281 in mouse embryonic stem (ES) cells. ZFP281 recruits AFF3 to the Meg3 enhancer within the imprinted Dlk1-Dio3 locus, thus regulating the allele-specific expression of the Meg3 polycistron. Our genome-wide analyses further identify ZFP281 as a critical factor generally associating with AFF3 at enhancers and functioning together with AFF3 in regulating the expression of a subset of genes. Our study suggests that different zinc finger proteins can recruit AFF3 to different regulatory elements and differentially regulate the function of AFF3 in a context-dependent manner.
Figures

References
-
- Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 2014; 15:517–530. - PubMed
-
- Lee J.T., Bartolomei M.S.. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013; 152:1308–1323. - PubMed
-
- Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A. et al. . In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011; 44:361–372. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

