RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes
- PMID: 28180304
- PMCID: PMC5605241
- DOI: 10.1093/nar/gkw1239
RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes
Abstract
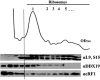
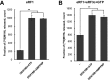
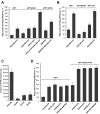
The human DEAD-box RNA-helicase DDX19 functions in mRNA export through the nuclear pore complex. The yeast homolog of this protein, Dbp5, has been reported to participate in translation termination. Using a reconstituted mammalian in vitro translation system, we show that the human protein DDX19 is also important for translation termination. It is associated with the fraction of translating ribosomes. We show that DDX19 interacts with pre-termination complexes (preTCs) in a nucleotide-dependent manner. Furthermore, DDX19 increases the efficiency of termination complex (TC) formation and the peptide release in the presence of eukaryotic release factors. Using the eRF1(AGQ) mutant protein or a non-hydrolysable analog of GTP to inhibit subsequent peptidyl-tRNA hydrolysis, we reveal that the activation of translation termination by DDX19 occurs during the stop codon recognition. This activation is a result of DDX19 binding to preTC and a concomitant stabilization of terminating ribosomes. Moreover, we show that DDX19 stabilizes ribosome complexes with translation elongation factors eEF1 and eEF2. Taken together, our findings reveal that the human RNA helicase DDX19 actively participates in protein biosynthesis.
Figures



Similar articles
-
Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay.Int J Mol Sci. 2020 Feb 6;21(3):1085. doi: 10.3390/ijms21031085. Int J Mol Sci. 2020. PMID: 32041247 Free PMC article. Review.
-
The DEAD-box RNA helicase Dbp5 functions in translation termination.Science. 2007 Feb 2;315(5812):646-9. doi: 10.1126/science.1134641. Science. 2007. PMID: 17272721
-
Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3.Nucleic Acids Res. 2019 May 21;47(9):4798-4813. doi: 10.1093/nar/gkz177. Nucleic Acids Res. 2019. PMID: 30873535 Free PMC article.
-
Translation termination: new factors and insights.RNA Biol. 2010 Sep-Oct;7(5):548-50. doi: 10.4161/rna.7.5.12686. Epub 2010 Sep 1. RNA Biol. 2010. PMID: 21081843
-
Dbp5 - from nuclear export to translation.Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
-
Chemical genetic inhibition of DEAD-box proteins using covalent complementarity.Nucleic Acids Res. 2018 Sep 28;46(17):8689-8699. doi: 10.1093/nar/gky706. Nucleic Acids Res. 2018. PMID: 30102385 Free PMC article.
-
DEAD-ly Affairs: The Roles of DEAD-Box Proteins on HIV-1 Viral RNA Metabolism.Front Cell Dev Biol. 2022 Jun 13;10:917599. doi: 10.3389/fcell.2022.917599. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35769258 Free PMC article. Review.
-
Elongation factor 2 in cancer: a promising therapeutic target in protein translation.Cell Mol Biol Lett. 2024 Dec 20;29(1):156. doi: 10.1186/s11658-024-00674-7. Cell Mol Biol Lett. 2024. PMID: 39707196 Free PMC article. Review.
-
CTELS: A Cell-Free System for the Analysis of Translation Termination Rate.Biomolecules. 2020 Jun 16;10(6):911. doi: 10.3390/biom10060911. Biomolecules. 2020. PMID: 32560154 Free PMC article.
-
Nonsense suppression therapies in human genetic diseases.Cell Mol Life Sci. 2021 May;78(10):4677-4701. doi: 10.1007/s00018-021-03809-7. Epub 2021 Mar 22. Cell Mol Life Sci. 2021. PMID: 33751142 Free PMC article. Review.
References
-
- Linder P., Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. - PubMed
-
- Linder P., Fuller-Pace F.V.. Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta. 2013; 1829:750–755. - PubMed
-
- Cordin O., Banroques J., Tanner N.K., Linder P.. The DEAD-box protein family of RNA helicases. Gene. 2006; 367:17–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

