Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae
- PMID: 28180315
- PMCID: PMC5388407
- DOI: 10.1093/nar/gkw1224
Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae
Abstract
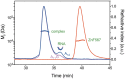
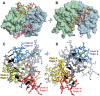
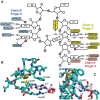
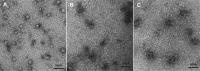
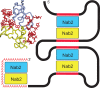
In Saccharomyces cerevisiae generation of export-competent mRNPs terminates the nuclear phase of the gene expression pathway and facilitates transport to the cytoplasm for translation. Nab2 functions in this process to control both mRNP compaction that facilitates movement through nuclear pore complexes and the length of transcript poly(A) tails. Nab2 has a modular structure that includes seven CCCH Zn fingers that bind to A-rich RNAs and fingers 5–7 are critical for these functions. Here, we demonstrate, using both biophysical and structural methods, that binding A11G RNA induces dimerization of Zn fingers 5–7 mediated by the novel spatial arrangement of the fingers promoting each RNA chain binding two protein chains. The dimerization of Nab2 induced by RNA binding provides a basis for understanding its function in both poly(A) tail length regulation and in the compaction of mature transcripts to facilitate nuclear export.
Figures
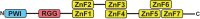

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

