Analysis of translation using polysome profiling
- PMID: 28180329
- PMCID: PMC5388431
- DOI: 10.1093/nar/gkw907
Analysis of translation using polysome profiling
Abstract
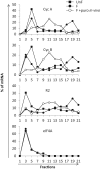
During the past decade, there has been growing interest in the role of translational regulation of gene expression in many organisms. Polysome profiling has been developed to infer the translational status of a specific mRNA species or to analyze the translatome, i.e. the subset of mRNAs actively translated in a cell. Polysome profiling is especially suitable for emergent model organisms for which genomic data are limited. In this paper, we describe an optimized protocol for the purification of sea urchin polysomes and highlight the critical steps involved in polysome purification. We applied this protocol to obtain experimental results on translational regulation of mRNAs following fertilization. Our protocol should prove useful for integrating the study of the role of translational regulation in gene regulatory networks in any biological model. In addition, we demonstrate how to carry out high-throughput processing of polysome gradient fractions, for the simultaneous screening of multiple biological conditions and large-scale preparation of samples for next-generation sequencing.
Figures



References
-
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

