PARP-1/PARP-2 double deficiency in mouse T cells results in faulty immune responses and T lymphomas
- PMID: 28181505
- PMCID: PMC5299517
- DOI: 10.1038/srep41962
PARP-1/PARP-2 double deficiency in mouse T cells results in faulty immune responses and T lymphomas
Abstract
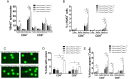
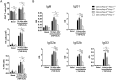
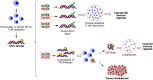
The maintenance of T-cell homeostasis must be tightly regulated. Here, we have identified a coordinated role of Poly(ADP-ribose) polymerase-1 (PARP-1) and PARP-2 in maintaining T-lymphocyte number and function. Mice bearing a T-cell specific deficiency of PARP-2 in a PARP-1-deficient background showed defective thymocyte maturation and diminished numbers of peripheral CD4+ and CD8+ T-cells. Meanwhile, peripheral T-cell number was not affected in single PARP-1 or PARP-2-deficient mice. T-cell lymphopenia was associated with dampened in vivo immune responses to synthetic T-dependent antigens and virus, increased DNA damage and T-cell death. Moreover, double-deficiency in PARP-1/PARP-2 in T-cells led to highly aggressive T-cell lymphomas with long latency. Our findings establish a coordinated role of PARP-1 and PARP-2 in T-cell homeostasis that might impact on the development of PARP-centred therapies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Rothenberg E. V. & Taghon T. Molecular genetics of T cell development. Annu. Rev. Immunol. 23, 601–649 (2005). - PubMed
-
- Grossman Z. & Paul W. E. Dynamic tuning of lymphocytes: physiological basis, mechanisms, and function. Annu. Rev. Immunol. 33, 677–713 (2015). - PubMed
-
- Surh C. D. & Sprent J. Homeostasis of naive and memory T cells. Immunity. 29, 848–862 (2008). - PubMed
-
- Baek K. H. et al.. p53 deficiency and defective mitotic checkpoint in proliferating T lymphocytes increase chromosomal instability through aberrant exit from mitotic arrest. J. Leukoc. Biol. 73, 850–861 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

