Tenovin-6 impairs autophagy by inhibiting autophagic flux
- PMID: 28182004
- PMCID: PMC5386474
- DOI: 10.1038/cddis.2017.25
Tenovin-6 impairs autophagy by inhibiting autophagic flux
Erratum in
-
Correction: Tenovin-6 impairs autophagy by inhibiting autophagic flux.Cell Death Dis. 2018 Jul 16;9(8):790. doi: 10.1038/s41419-018-0826-0. Cell Death Dis. 2018. PMID: 30013038 Free PMC article.
Abstract
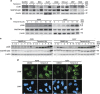
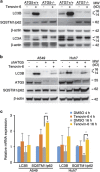
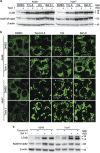
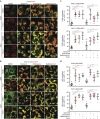
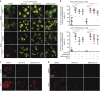
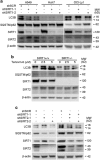
Tenovin-6 has attracted significant interest because it activates p53 and inhibits sirtuins. It has anti-neoplastic effects on multiple hematopoietic malignancies and solid tumors in both in vitro and in vivo studies. Tenovin-6 was recently shown to impair the autophagy pathway in chronic lymphocytic leukemia cells and pediatric soft tissue sarcoma cells. However, whether tenovin-6 has a general inhibitory effect on autophagy and whether there is any involvement with SIRT1 and p53, both of which are regulators of the autophagy pathway, remain unclear. In this study, we have demonstrated that tenovin-6 increases microtubule-associated protein 1 light chain 3 (LC3-II) level in diverse cell types in a time- and dose-dependent manner. Mechanistically, the increase of LC3-II by tenovin-6 is caused by inhibition of the classical autophagy pathway via impairing lysosomal function without affecting the fusion between autophagosomes and lysosomes. Furthermore, we have revealed that tenovin-6 activation of p53 is cell type dependent, and tenovin-6 inhibition of autophagy is not dependent on its regulatory functions on p53 and SIRT1. Our results have shown that tenovin-6 is a potent autophagy inhibitor, and raised the precaution in interpreting results where tenovin-6 is used as an inhibitor of SIRT1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Tenovin-6 inhibits proliferation and survival of diffuse large B-cell lymphoma cells by blocking autophagy.Oncotarget. 2017 Feb 28;8(9):14912-14924. doi: 10.18632/oncotarget.14741. Oncotarget. 2017. PMID: 28118604 Free PMC article.
-
Dysregulation of autophagy in chronic lymphocytic leukemia with the small-molecule Sirtuin inhibitor Tenovin-6.Sci Rep. 2013;3:1275. doi: 10.1038/srep01275. Sci Rep. 2013. PMID: 23429453 Free PMC article.
-
SIRT1 and SIRT2 inhibition impairs pediatric soft tissue sarcoma growth.Cell Death Dis. 2014 Oct 23;5(10):e1483. doi: 10.1038/cddis.2014.385. Cell Death Dis. 2014. PMID: 25341037 Free PMC article.
-
SIRT1 - a new mammalian substrate of nuclear autophagy.Autophagy. 2021 Feb;17(2):593-595. doi: 10.1080/15548627.2020.1860541. Epub 2020 Dec 20. Autophagy. 2021. PMID: 33292048 Free PMC article. Review.
-
How to interpret LC3 immunoblotting.Autophagy. 2007 Nov-Dec;3(6):542-5. doi: 10.4161/auto.4600. Epub 2007 Jun 19. Autophagy. 2007. PMID: 17611390 Review.
Cited by
-
Heterogeneous Responses of Gastric Cancer Cell Lines to Tenovin-6 and Synergistic Effect with Chloroquine.Cancers (Basel). 2020 Feb 5;12(2):365. doi: 10.3390/cancers12020365. Cancers (Basel). 2020. PMID: 32033497 Free PMC article.
-
The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy.Front Cell Dev Biol. 2020 Sep 29;8:576946. doi: 10.3389/fcell.2020.576946. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33117804 Free PMC article. Review.
-
Determinants of Ion-Transporter Cancer Cell Death.Chem. 2019 Aug 8;5(8):2079-2098. doi: 10.1016/j.chempr.2019.05.001. Epub 2019 May 20. Chem. 2019. PMID: 33791443 Free PMC article.
-
CRISPR-Cas9 Screening of Kaposi's Sarcoma-Associated Herpesvirus-Transformed Cells Identifies XPO1 as a Vulnerable Target of Cancer Cells.mBio. 2019 May 14;10(3):e00866-19. doi: 10.1128/mBio.00866-19. mBio. 2019. PMID: 31088931 Free PMC article.
-
Therapeutic Modulation of Autophagy in Leukaemia and Lymphoma.Cells. 2019 Jan 30;8(2):103. doi: 10.3390/cells8020103. Cells. 2019. PMID: 30704144 Free PMC article. Review.
References
-
- McCarthy AR, Pirrie L, Hollick JJ, Ronseaux S, Campbell J, Higgins M et al. Synthesis and biological characterisation of sirtuin inhibitors based on the tenovins. Bioorg Med Chem 2012; 20: 1779–1793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

