Prenatal alcohol exposure impairs autophagy in neonatal brain cortical microvessels
- PMID: 28182007
- PMCID: PMC5386476
- DOI: 10.1038/cddis.2017.29
Prenatal alcohol exposure impairs autophagy in neonatal brain cortical microvessels
Abstract
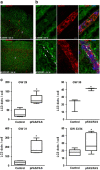
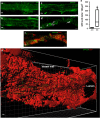
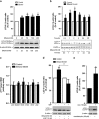
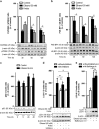
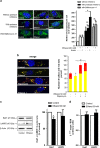
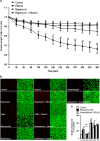
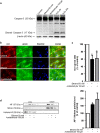
Brain developmental lesions are a devastating consequence of prenatal alcohol exposure (PAE). We recently showed that PAE affects cortical vascular development with major effects on angiogenesis and endothelial cell survival. The underlying molecular mechanisms of these effects remain poorly understood. This study aimed at characterizing the ethanol exposure impact on the autophagic process in brain microvessels in human fetuses with fetal alcohol syndrome (FAS) and in a PAE mouse model. Our results indicate that PAE induces an increase of autophagic vacuole number in human fetal and neonatal mouse brain cortical microvessels. Subsequently, ex vivo studies using green fluorescent protein (GFP)-LC3 mouse microvessel preparations revealed that ethanol treatment alters autophagy in endothelial cells. Primary cultures of mouse brain microvascular endothelial cells were used to characterize the underlying molecular mechanisms. LC3 and p62 protein levels were significantly increased in endothelial cells treated with 50 mM ethanol. The increase of autophagic vacuole number may be due to excessive autophagosome formation associated with the partial inhibition of the mammalian target of rapamycin pathway upon ethanol exposure. In addition, the progression from autophagosomes to autolysosomes, which was monitored using autophagic flux inhibitors and mRFP-EGFP vector, showed a decrease in the autolysosome number. Besides, a decrease in the Rab7 protein level was observed that may underlie the impairment of autophagosome-lysosome fusion. In addition, our results showed that ethanol-induced cell death is likely to be mediated by decreased mitochondrial integrity and release of apoptosis-inducing factor. Interestingly, incubation of cultured cells with rapamycin prevented ethanol effects on autophagic flux, ethanol-induced cell death and vascular plasticity. Taken together, these results are consistent with autophagy dysregulation in cortical microvessels upon ethanol exposure, which could contribute to the defects in angiogenesis observed in patients with FAS. Moreover, our results suggest that rapamycin represents a potential therapeutic strategy to reduce PAE-related brain developmental disorders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Archibald SL, Fennema-Notestine C, Gamst A, Riley EP, Mattson SN, Jernigan TL. Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev Med Child Neurol 2001; 43: 148–154. - PubMed
-
- Burd L, Martsolf JT, Klug MG, Kerbeshian J. Diagnosis of FAS: a comparison of the Fetal Alcohol Syndrome Diagnostic Checklist and the Institute of Medicine Criteria for Fetal Alcohol Syndrome. Neurotoxicol Teratol 2003; 25: 719–724. - PubMed
-
- Ward RJ, Colivicchi MA, Allen R, Schol F, Lallemand F, de Witte P et al. Neuro-inflammation induced in the hippocampus of 'binge drinking' rats may be mediated by elevated extracellular glutamate content. J Neurochem 2009; 111: 1119–1128. - PubMed
-
- Maier SE, Miller JA, West JR. Prenatal binge-like alcohol exposure in the rat results in region-specific deficits in brain growth. Neurotoxicol Teratol 1999; 21: 285–291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

