TAK1 regulates resident macrophages by protecting lysosomal integrity
- PMID: 28182011
- PMCID: PMC5386472
- DOI: 10.1038/cddis.2017.23
TAK1 regulates resident macrophages by protecting lysosomal integrity
Abstract
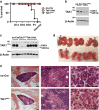
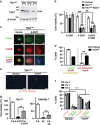
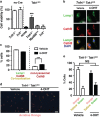
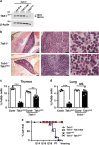
Hematopoietic cell survival and death is critical for development of a functional immune system. Here, we report that a protein kinase, TAK1, is selectively required for resident macrophage integrity during embryogenesis. Hematopoietic lineage-specific deletion of Tak1 gene (Tak1HKO) caused accumulation of cellular debris in the thymus in perinatal mice. Although no overt alteration in thymocytes and blood myeloid populations was observed in Tak1HKO mice, we found that thymic and lung macrophages were diminished. In the in vitro setting, Tak1 deficiency caused profound disruption of lysosomes and killed bone marrow-derived macrophages (BMDMs) without any exogenous stressors. Inhibition of the lysosomal protease, cathepsin B, partially blocked Tak1-deficient BMDM death, suggesting that leakage of the lysosomal contents is in part the cause of cell death. To identify the trigger of this cell death, we examined involvement of TNF and Toll-like receptor pathways. Among them, we found that deletion of Tnfr1 partially rescued cell death. Finally, we show that Tnfr1 deletion partially restored thymic and lung macrophages in vivo. These results suggest that autocrine and potentially paracrine TNF kills Tak1-deficient macrophages during development. Our results reveal that TAK1 signaling maintains proper macrophage populations through protecting lysosomal integrity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 2016; 44: 439–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

