RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides
- PMID: 28182648
- PMCID: PMC5325595
- DOI: 10.1371/journal.pgen.1006613
RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides
Abstract
Locomotor activity rhythms are controlled by a network of ~150 circadian neurons within the adult Drosophila brain. They are subdivided based on their anatomical locations and properties. We profiled transcripts "around the clock" from three key groups of circadian neurons with different functions. We also profiled a non-circadian outgroup, dopaminergic (TH) neurons. They have cycling transcripts but fewer than clock neurons as well as low expression and poor cycling of clock gene transcripts. This suggests that TH neurons do not have a canonical circadian clock and that their gene expression cycling is driven by brain systemic cues. The three circadian groups are surprisingly diverse in their cycling transcripts and overall gene expression patterns, which include known and putative novel neuropeptides. Even the overall phase distributions of cycling transcripts are distinct, indicating that different regulatory principles govern transcript oscillations. This surprising cell-type diversity parallels the functional heterogeneity of the different neurons.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
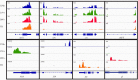

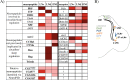

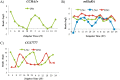
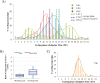
References
-
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99(7):791–802. Epub 2000/01/05. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

