Representing high throughput expression profiles via perturbation barcodes reveals compound targets
- PMID: 28182661
- PMCID: PMC5300121
- DOI: 10.1371/journal.pcbi.1005335
Representing high throughput expression profiles via perturbation barcodes reveals compound targets
Abstract
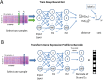
High throughput mRNA expression profiling can be used to characterize the response of cell culture models to perturbations such as pharmacologic modulators and genetic perturbations. As profiling campaigns expand in scope, it is important to homogenize, summarize, and analyze the resulting data in a manner that captures significant biological signals in spite of various noise sources such as batch effects and stochastic variation. We used the L1000 platform for large-scale profiling of 978 representative genes across thousands of compound treatments. Here, a method is described that uses deep learning techniques to convert the expression changes of the landmark genes into a perturbation barcode that reveals important features of the underlying data, performing better than the raw data in revealing important biological insights. The barcode captures compound structure and target information, and predicts a compound's high throughput screening promiscuity, to a higher degree than the original data measurements, indicating that the approach uncovers underlying factors of the expression data that are otherwise entangled or masked by noise. Furthermore, we demonstrate that visualizations derived from the perturbation barcode can be used to more sensitively assign functions to unknown compounds through a guilt-by-association approach, which we use to predict and experimentally validate the activity of compounds on the MAPK pathway. The demonstrated application of deep metric learning to large-scale chemical genetics projects highlights the utility of this and related approaches to the extraction of insights and testable hypotheses from big, sometimes noisy data.
Conflict of interest statement
At the time of submission, all of the authors were paid employees of Merck & Co., Inc. (Kenilworth, NJ).
Figures


References
-
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science (New York, NY. 2006;313(5795):1929–35. - PubMed
-
- Waring JF, Ciurlionis R, Jolly RA, Heindel M, Ulrich RG. Microarray analysis of hepatotoxins in vitro reveals a correlation between gene expression profiles and mechanisms of toxicity. Toxicol Lett. 2001;120(1–3):359–68. - PubMed
-
- Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14621–6. 10.1073/pnas.1000138107 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

