Streptomyces aridus sp. nov., isolated from a high altitude Atacama Desert soil and emended description of Streptomyces noboritoensis Isono et al. 1957
- PMID: 28185026
- PMCID: PMC5387016
- DOI: 10.1007/s10482-017-0838-2
Streptomyces aridus sp. nov., isolated from a high altitude Atacama Desert soil and emended description of Streptomyces noboritoensis Isono et al. 1957
Abstract
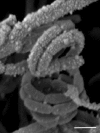
A polyphasic study was undertaken to determine the taxonomic status of a Streptomyces strain which had been isolated from a high altitude Atacama Desert soil and shown to have bioactive properties. The strain, isolate H9T, was found to have chemotaxonomic, cultural and morphological properties that place it in the genus Streptomyces. 16S rRNA gene sequence analyses showed that the isolate forms a distinct branch at the periphery of a well-delineated subclade in the Streptomyces 16S rRNA gene tree together with the type strains of Streptomyces crystallinus, Streptomyces melanogenes and Streptomyces noboritoensis. Multi-locus sequence analysis (MLSA) based on five house-keeping gene alleles showed that isolate H9T is closely related to the latter two type strains and to Streptomyces polyantibioticus NRRL B-24448T. The isolate was distinguished readily from the type strains of S. melanogenes, S. noboritoensis and S. polyantibioticus using a combination of phenotypic properties. Consequently, the isolate is considered to represent a new species of Streptomyces for which the name Streptomyces aridus sp. nov. is proposed; the type strain is H9T (=NCIMB 14965T=NRRL B65268T). In addition, the MLSA and phenotypic data show that the S. melanogenes and S. noboritoensis type strains belong to a single species, it is proposed that S. melanogenes be recognised as a heterotypic synonym of S. noboritoensis for which an emended description is given.
Keywords: Aridus; Atacama Desert; Polyphasic taxonomy; Streptomyces.
Figures


References
-
- Athalye M, Lacey J, Goodfellow M. Selective isolation and enumeration of actinomycetes using rifampicin. J Appl Bacteriol. 1981;51:289–297. doi: 10.1111/j.1365-2672.1981.tb01244.x. - DOI
-
- Bull AT. Actinobacteria of the extremobiosphere. In: Horikoshi K, editor. Extremophiles Handbook. Tokyo: Springer; 2011. pp. 1203–1240.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

