P113 is a merozoite surface protein that binds the N terminus of Plasmodium falciparum RH5
- PMID: 28186186
- PMCID: PMC5309799
- DOI: 10.1038/ncomms14333
P113 is a merozoite surface protein that binds the N terminus of Plasmodium falciparum RH5
Abstract
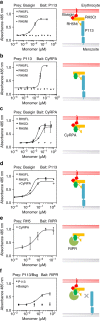
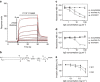
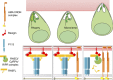
Invasion of erythrocytes by Plasmodium falciparum merozoites is necessary for malaria pathogenesis and is therefore a primary target for vaccine development. RH5 is a leading subunit vaccine candidate because anti-RH5 antibodies inhibit parasite growth and the interaction with its erythrocyte receptor basigin is essential for invasion. RH5 is secreted, complexes with other parasite proteins including CyRPA and RIPR, and contains a conserved N-terminal region (RH5Nt) of unknown function that is cleaved from the native protein. Here, we identify P113 as a merozoite surface protein that directly interacts with RH5Nt. Using recombinant proteins and a sensitive protein interaction assay, we establish the binding interdependencies of all the other known RH5 complex components and conclude that the RH5Nt-P113 interaction provides a releasable mechanism for anchoring RH5 to the merozoite surface. We exploit these findings to design a chemically synthesized peptide corresponding to RH5Nt, which could contribute to a cost-effective malaria vaccine.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
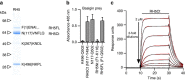
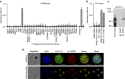
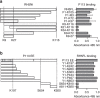
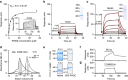
References
-
- Murray C. J. et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379, 413–431 (2012). - PubMed
-
- Cowman A. F. & Crabb B. S. Invasion of red blood cells by malaria parasites. Cell 124, 755–766 (2006). - PubMed
-
- Hadley T. J. Invasion of erythrocytes by malaria parasites: a cellular and molecular overview. Annu. Rev. Microbiol. 40, 451–477 (1986). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

