An Atlas of Peroxiredoxins Created Using an Active Site Profile-Based Approach to Functionally Relevant Clustering of Proteins
- PMID: 28187133
- PMCID: PMC5302317
- DOI: 10.1371/journal.pcbi.1005284
An Atlas of Peroxiredoxins Created Using an Active Site Profile-Based Approach to Functionally Relevant Clustering of Proteins
Abstract
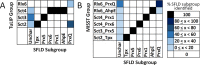
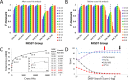
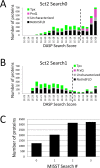
Peroxiredoxins (Prxs or Prdxs) are a large protein superfamily of antioxidant enzymes that rapidly detoxify damaging peroxides and/or affect signal transduction and, thus, have roles in proliferation, differentiation, and apoptosis. Prx superfamily members are widespread across phylogeny and multiple methods have been developed to classify them. Here we present an updated atlas of the Prx superfamily identified using a novel method called MISST (Multi-level Iterative Sequence Searching Technique). MISST is an iterative search process developed to be both agglomerative, to add sequences containing similar functional site features, and divisive, to split groups when functional site features suggest distinct functionally-relevant clusters. Superfamily members need not be identified initially-MISST begins with a minimal representative set of known structures and searches GenBank iteratively. Further, the method's novelty lies in the manner in which isofunctional groups are selected; rather than use a single or shifting threshold to identify clusters, the groups are deemed isofunctional when they pass a self-identification criterion, such that the group identifies itself and nothing else in a search of GenBank. The method was preliminarily validated on the Prxs, as the Prxs presented challenges of both agglomeration and division. For example, previous sequence analysis clustered the Prx functional families Prx1 and Prx6 into one group. Subsequent expert analysis clearly identified Prx6 as a distinct functionally relevant group. The MISST process distinguishes these two closely related, though functionally distinct, families. Through MISST search iterations, over 38,000 Prx sequences were identified, which the method divided into six isofunctional clusters, consistent with previous expert analysis. The results represent the most complete computational functional analysis of proteins comprising the Prx superfamily. The feasibility of this novel method is demonstrated by the Prx superfamily results, laying the foundation for potential functionally relevant clustering of the universe of protein sequences.
Conflict of interest statement
The authors have declared no competing interests exist.
Figures








References
-
- Poole LB. The catalytic mechanism of peroxiredoxins In: Flohé L, Harris JR, editors. Peroxiredoxin systems. New York: Springer; 2007. pp. 61–81.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

