Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer
- PMID: 28188128
- PMCID: PMC5460679
- DOI: 10.1158/2159-8290.CD-16-1045
Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer
Abstract
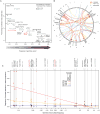
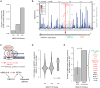
Recent studies have characterized the extensive somatic alterations that arise during cancer. However, the somatic evolution of a tumor may be significantly affected by inherited polymorphisms carried in the germline. Here, we analyze genomic data for 5,954 tumors to reveal and systematically validate 412 genetic interactions between germline polymorphisms and major somatic events, including tumor formation in specific tissues and alteration of specific cancer genes. Among germline-somatic interactions, we found germline variants in RBFOX1 that increased incidence of SF3B1 somatic mutation by 8-fold via functional alterations in RNA splicing. Similarly, 19p13.3 variants were associated with a 4-fold increased likelihood of somatic mutations in PTEN. In support of this association, we found that PTEN knockdown sensitizes the MTOR pathway to high expression of the 19p13.3 gene GNA11 Finally, we observed that stratifying patients by germline polymorphisms exposed distinct somatic mutation landscapes, implicating new cancer genes. This study creates a validated resource of inherited variants that govern where and how cancer develops, opening avenues for prevention research.Significance: This study systematically identifies germline variants that directly affect tumor evolution, either by dramatically increasing alteration frequency of specific cancer genes or by influencing the site where a tumor develops. Cancer Discovery; 7(4); 410-23. ©2017 AACR.See related commentary by Geeleher and Huang, p. 354This article is highlighted in the In This Issue feature, p. 339.
©2017 American Association for Cancer Research.
Conflict of interest statement
COI Disclosure Statement: The authors declare no potential conflicts of interest.
Figures




Comment in
-
Genomics: A different view of the landscape.Nat Rev Cancer. 2017 Apr;17(4):206-207. doi: 10.1038/nrc.2017.22. Epub 2017 Mar 10. Nat Rev Cancer. 2017. PMID: 28280266 No abstract available.
-
Exploring the Link between the Germline and Somatic Genome in Cancer.Cancer Discov. 2017 Apr;7(4):354-355. doi: 10.1158/2159-8290.CD-17-0192. Cancer Discov. 2017. PMID: 28373166 Free PMC article.
References
-
- Collins FS, Barker AD. Mapping the cancer genome Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296(3):50–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

