An improved FSL-FIRST pipeline for subcortical gray matter segmentation to study abnormal brain anatomy using quantitative susceptibility mapping (QSM)
- PMID: 28188873
- PMCID: PMC5421359
- DOI: 10.1016/j.mri.2017.02.002
An improved FSL-FIRST pipeline for subcortical gray matter segmentation to study abnormal brain anatomy using quantitative susceptibility mapping (QSM)
Abstract
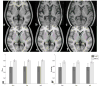
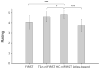
Accurate and robust segmentation of subcortical gray matter (SGM) nuclei is required in many neuroimaging applications. FMRIB's Integrated Registration and Segmentation Tool (FIRST) is one of the most popular software tools for automated subcortical segmentation based on T1-weighted (T1w) images. In this work, we demonstrate that FIRST tends to produce inaccurate SGM segmentation results in the case of abnormal brain anatomy, such as present in atrophied brains, due to a poor spatial match of the subcortical structures with the training data in the MNI space as well as due to insufficient contrast of SGM structures on T1w images. Consequently, such deviations from the average brain anatomy may introduce analysis bias in clinical studies, which may not always be obvious and potentially remain unidentified. To improve the segmentation of subcortical nuclei, we propose to use FIRST in combination with a special Hybrid image Contrast (HC) and Non-Linear (nl) registration module (HC-nlFIRST), where the hybrid image contrast is derived from T1w images and magnetic susceptibility maps to create subcortical contrast that is similar to that in the Montreal Neurological Institute (MNI) template. In our approach, a nonlinear registration replaces FIRST's default linear registration, yielding a more accurate alignment of the input data to the MNI template. We evaluated our method on 82 subjects with particularly abnormal brain anatomy, selected from a database of >2000 clinical cases. Qualitative and quantitative analyses revealed that HC-nlFIRST provides improved segmentation compared to the default FIRST method.
Keywords: Brain; FIRST; MRI; Quantitative susceptibility mapping; Segmentation; Subcortical gray matter.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures









Similar articles
-
Multi-atlas tool for automated segmentation of brain gray matter nuclei and quantification of their magnetic susceptibility.Neuroimage. 2019 May 1;191:337-349. doi: 10.1016/j.neuroimage.2019.02.016. Epub 2019 Feb 7. Neuroimage. 2019. PMID: 30738207 Free PMC article.
-
Human brain atlas for automated region of interest selection in quantitative susceptibility mapping: application to determine iron content in deep gray matter structures.Neuroimage. 2013 Nov 15;82:449-69. doi: 10.1016/j.neuroimage.2013.05.127. Epub 2013 Jun 12. Neuroimage. 2013. PMID: 23769915 Free PMC article.
-
Subcortical gray matter segmentation and voxel-based analysis using transverse relaxation and quantitative susceptibility mapping with application to multiple sclerosis.J Magn Reson Imaging. 2015 Dec;42(6):1601-10. doi: 10.1002/jmri.24951. Epub 2015 May 18. J Magn Reson Imaging. 2015. PMID: 25980643
-
Quantifying deep grey matter atrophy using automated segmentation approaches: A systematic review of structural MRI studies.Neuroimage. 2019 Nov 1;201:116018. doi: 10.1016/j.neuroimage.2019.116018. Epub 2019 Jul 15. Neuroimage. 2019. PMID: 31319182
-
[Understanding Voxel-Based Morphometry].Brain Nerve. 2017 May;69(5):505-511. doi: 10.11477/mf.1416200776. Brain Nerve. 2017. PMID: 28479527 Review. Japanese.
Cited by
-
DBSegment: Fast and robust segmentation of deep brain structures considering domain generalization.Hum Brain Mapp. 2023 Feb 1;44(2):762-778. doi: 10.1002/hbm.26097. Epub 2022 Oct 17. Hum Brain Mapp. 2023. PMID: 36250712 Free PMC article.
-
Accuracy of automated amygdala MRI segmentation approaches in Huntington's disease in the IMAGE-HD cohort.Hum Brain Mapp. 2020 May;41(7):1875-1888. doi: 10.1002/hbm.24918. Epub 2020 Feb 7. Hum Brain Mapp. 2020. PMID: 32034838 Free PMC article.
-
Decreasing brain iron in multiple sclerosis: The difference between concentration and content in iron MRI.Hum Brain Mapp. 2021 Apr 1;42(5):1463-1474. doi: 10.1002/hbm.25306. Epub 2020 Dec 30. Hum Brain Mapp. 2021. PMID: 33378095 Free PMC article.
-
Implicit associative learning relates to basal ganglia gray matter microstructure in young and older adults.Behav Brain Res. 2021 Jan 15;397:112950. doi: 10.1016/j.bbr.2020.112950. Epub 2020 Oct 2. Behav Brain Res. 2021. PMID: 33017642 Free PMC article.
-
Convergent imaging-transcriptomic evidence for disturbed iron homeostasis in Gilles de la Tourette syndrome.medRxiv [Preprint]. 2023 May 16:2023.05.15.23289978. doi: 10.1101/2023.05.15.23289978. medRxiv. 2023. Update in: Neurobiol Dis. 2023 Sep;185:106252. doi: 10.1016/j.nbd.2023.106252. PMID: 37292704 Free PMC article. Updated. Preprint.
References
-
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–55. - PubMed
-
- Fischl B, Salat DH, van der Kouwe AJ, Makris N, Segonne F, Quinn BT, et al. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23(Suppl 1):69–84. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

