CD8+ T Cells Orchestrate pDC-XCR1+ Dendritic Cell Spatial and Functional Cooperativity to Optimize Priming
- PMID: 28190711
- PMCID: PMC5362251
- DOI: 10.1016/j.immuni.2017.01.003
CD8+ T Cells Orchestrate pDC-XCR1+ Dendritic Cell Spatial and Functional Cooperativity to Optimize Priming
Abstract
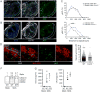
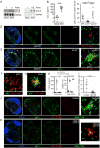
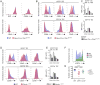
Adaptive cellular immunity is initiated by antigen-specific interactions between T lymphocytes and dendritic cells (DCs). Plasmacytoid DCs (pDCs) support antiviral immunity by linking innate and adaptive immune responses. Here we examined pDC spatiotemporal dynamics during viral infection to uncover when, where, and how they exert their functions. We found that pDCs accumulated at sites of CD8+ T cell antigen-driven activation in a CCR5-dependent fashion. Furthermore, activated CD8+ T cells orchestrated the local recruitment of lymph node-resident XCR1 chemokine receptor-expressing DCs via secretion of the XCL1 chemokine. Functionally, this CD8+ T cell-mediated reorganization of the local DC network allowed for the interaction and cooperation of pDCs and XCR1+ DCs, thereby optimizing XCR1+ DC maturation and cross-presentation. These data support a model in which CD8+ T cells upon activation create their own optimal priming microenvironment by recruiting additional DC subsets to the site of initial antigen recognition.
Keywords: CCL3; CCL4; CCR5; CXCR3; XCL1; chemokine; cooperation; migration; spatiotemporal; viral infection.
Published by Elsevier Inc.
Figures




Comment in
-
Antigen-Primed CD8+ T Cells Call DCs for Back Up.Immunity. 2017 Feb 21;46(2):163-164. doi: 10.1016/j.immuni.2017.02.002. Immunity. 2017. PMID: 28228271
References
-
- Ang DK, Oates CV, Schuelein R, Kelly M, Sansom FM, Bourges D, Boon L, Hertzog PJ, Hartland EL, van Driel IR. Cutting edge: pulmonary Legionella pneumophila is controlled by plasmacytoid dendritic cells but not type I IFN. J Immunol. 2010;184:5429–5433. - PubMed
-
- Asselin-Paturel C, Boonstra A, Dalod M, Durand I, Yessaad N, Dezutter-Dambuyant C, Vicari A, O’Garra A, Biron C, Briere F, et al. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat Immunol. 2001;2:1144–1150. - PubMed
-
- Barchet W, Krug A, Cella M, Newby C, Fischer JA, Dzionek A, Pekosz A, Colonna M. Dendritic cells respond to influenza virus through TLR7- and PKR-independent pathways. Eur J Immunol. 2005;35:236–242. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

