Serine ADP-Ribosylation Depends on HPF1
- PMID: 28190768
- PMCID: PMC5344681
- DOI: 10.1016/j.molcel.2017.01.003
Serine ADP-Ribosylation Depends on HPF1
Abstract
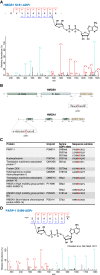
ADP-ribosylation (ADPr) regulates important patho-physiological processes through its attachment to different amino acids in proteins. Recently, by precision mapping on all possible amino acid residues, we identified histone serine ADPr marks in the DNA damage response. However, the biochemical basis underlying this serine modification remained unknown. Here we report that serine ADPr is strictly dependent on histone PARylation factor 1 (HPF1), a recently identified regulator of PARP-1. Quantitative proteomics revealed that serine ADPr does not occur in cells lacking HPF1. Moreover, adding HPF1 to in vitro PARP-1/PARP-2 reactions is necessary and sufficient for serine-specific ADPr of histones and PARP-1 itself. Three endogenous serine ADPr sites are located on the PARP-1 automodification domain. Further identification of serine ADPr on HMG proteins and hundreds of other targets indicates that serine ADPr is a widespread modification. We propose that O-linked protein ADPr is the key signal in PARP-1/PARP-2-dependent processes that govern genome stability.
Keywords: ADP-ribosylation; DNA damage; HPF1; PARP-1; PARP-2; genome stability; histones; proteomics; serine ADP-ribosylation.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
ADP-Ribosylation Goes Normal: Serine as the Major Site of the Modification.Cell Chem Biol. 2017 Apr 20;24(4):431-432. doi: 10.1016/j.chembiol.2017.04.003. Cell Chem Biol. 2017. PMID: 28431224
References
-
- Adamietz P. Poly(ADP-ribose) synthase is the major endogenous nonhistone acceptor for poly(ADP-ribose) in alkylated rat hepatoma cells. Eur. J. Biochem. 1987;169:365–372. - PubMed
-
- Bock F.J., Chang P. New directions in poly(ADP-ribose) polymerase biology. FEBS J. 2016;283:4017–4031. - PubMed
-
- Caldecott K.W. Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair (Amst.) 2014;19:108–113. - PubMed
-
- Chapman J.D., Gagné J.P., Poirier G.G., Goodlett D.R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2013;12:1868–1880. - PubMed
-
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

