OneDep: Unified wwPDB System for Deposition, Biocuration, and Validation of Macromolecular Structures in the PDB Archive
- PMID: 28190782
- PMCID: PMC5360273
- DOI: 10.1016/j.str.2017.01.004
OneDep: Unified wwPDB System for Deposition, Biocuration, and Validation of Macromolecular Structures in the PDB Archive
Abstract
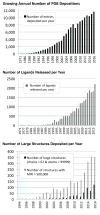
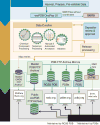
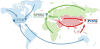
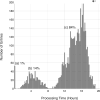
OneDep, a unified system for deposition, biocuration, and validation of experimentally determined structures of biological macromolecules to the PDB archive, has been developed as a global collaboration by the worldwide PDB (wwPDB) partners. This new system was designed to ensure that the wwPDB could meet the evolving archiving requirements of the scientific community over the coming decades. OneDep unifies deposition, biocuration, and validation pipelines across all wwPDB, EMDB, and BMRB deposition sites with improved focus on data quality and completeness in these archives, while supporting growth in the number of depositions and increases in their average size and complexity. In this paper, we describe the design, functional operation, and supporting infrastructure of the OneDep system, and provide initial performance assessments.
Keywords: 3D macromolecular structure; PDB; Protein Data Bank; biocuration; data archiving; data deposition; research data; structural biology; validation; wwPDB.
Published by Elsevier Ltd.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- BB/M013146/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM109046/GM/NIGMS NIH HHS/United States
- BB/J007471/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L007835/1/MRC_/Medical Research Council/United Kingdom
- 88944/WT_/Wellcome Trust/United Kingdom
- 75968/WT_/Wellcome Trust/United Kingdom
- BB/M020347/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K020013/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M020428/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM079429/GM/NIGMS NIH HHS/United States
- BB/M011674/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104948/WT_/Wellcome Trust/United Kingdom
- BB/K016970/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104948/WT_/Wellcome Trust/United Kingdom
- BB/G022577/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

