Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions
- PMID: 28191901
- PMCID: PMC5388574
- DOI: 10.1038/nbt.3803
Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions
Abstract
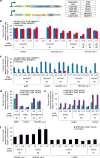
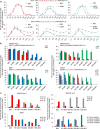
Base editing induces single-nucleotide changes in the DNA of living cells using a fusion protein containing a catalytically defective Streptococcus pyogenes Cas9, a cytidine deaminase, and an inhibitor of base excision repair. This genome editing approach has the advantage that it does not require formation of double-stranded DNA breaks or provision of a donor DNA template. Here we report the development of five C to T (or G to A) base editors that use natural and engineered Cas9 variants with different protospacer-adjacent motif (PAM) specificities to expand the number of sites that can be targeted by base editing 2.5-fold. Additionally, we engineered base editors containing mutated cytidine deaminase domains that narrow the width of the editing window from ∼5 nucleotides to as little as 1-2 nucleotides. We thereby enabled discrimination of neighboring C nucleotides, which would otherwise be edited with similar efficiency, and doubled the number of disease-associated target Cs able to be corrected preferentially over nearby non-target Cs.
Conflict of interest statement
The authors declare competing financial interests: D.R.L. is a consultant and co-founder of Editas Medicine, a company that seeks to develop genome-editing therapeutics. Y.B.K., A.C.K., and D.R.L. have filed patent applications on base editing.
Figures

Comment in
-
Base editing on the rise.Nat Biotechnol. 2017 May 9;35(5):428-429. doi: 10.1038/nbt.3871. Nat Biotechnol. 2017. PMID: 28486457 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

