Immune regulator ABIN1 suppresses HIV-1 transcription by negatively regulating the ubiquitination of Tat
- PMID: 28193275
- PMCID: PMC5304394
- DOI: 10.1186/s12977-017-0338-5
Immune regulator ABIN1 suppresses HIV-1 transcription by negatively regulating the ubiquitination of Tat
Abstract
Background: A20-binding inhibitor of NF-κB activation (ABIN1), an important immune regulator, was previously shown to be involved in HIV-1 replication. However, the reported studies done with overexpressed ABIN1 provided controversial results.
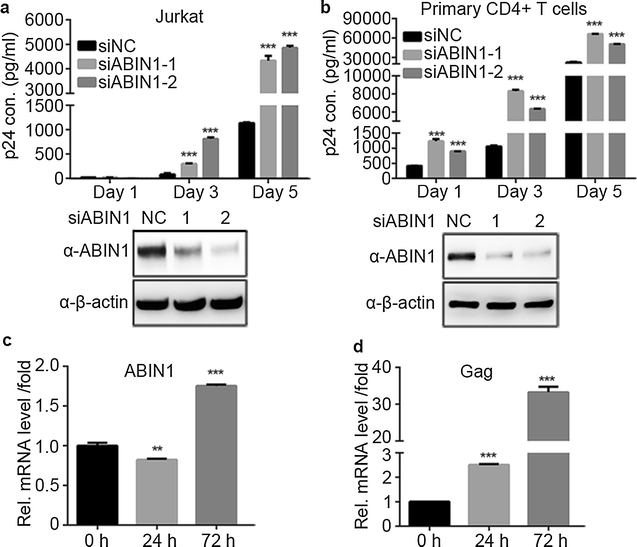
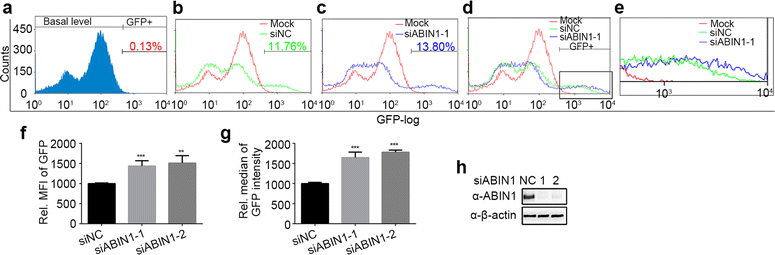
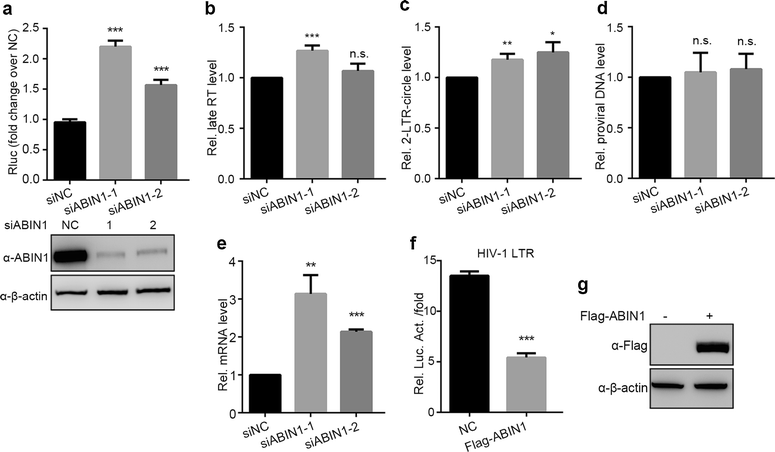
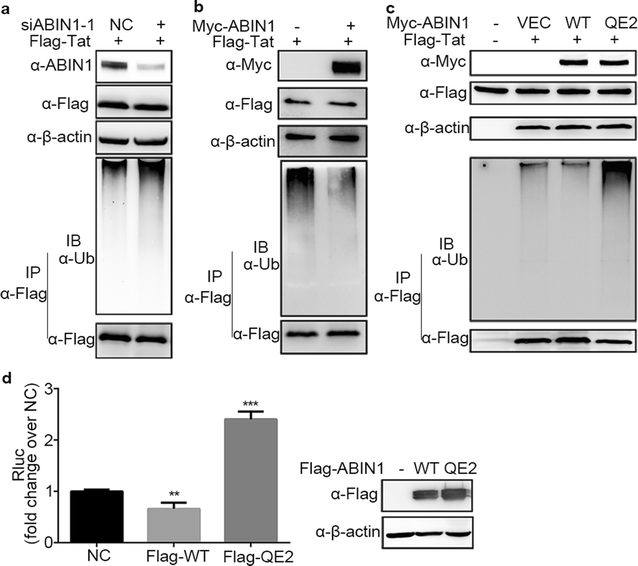
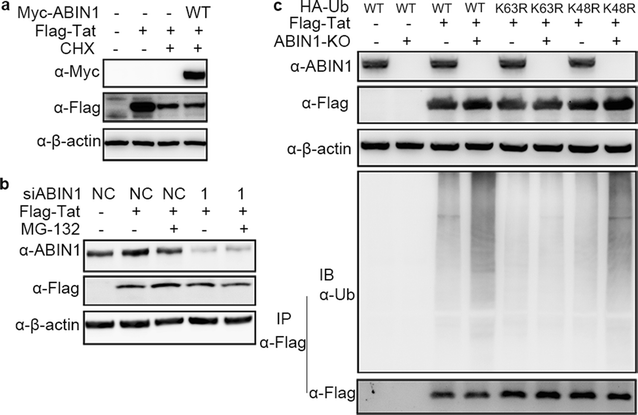
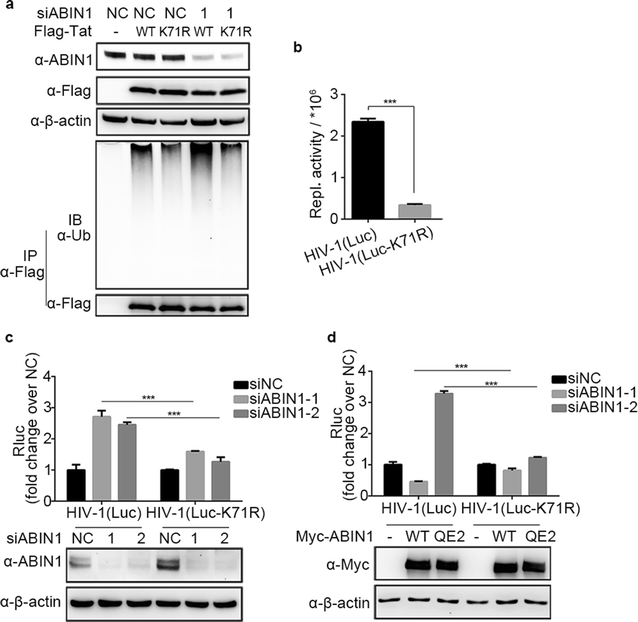
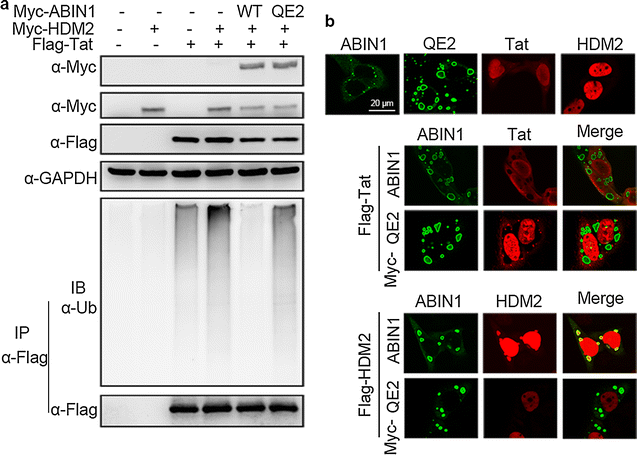
Results: Here we identified ABIN1 as a suppressor of HIV-1 transcription since transient knockdown of ABIN1 led to increased HIV-1 replication both in transformed Jurkat T cell line and in primary human CD4+ T lymphocytes. Depletion of ABIN1 specifically enhanced the HIV-1 transcription from the integrated genome during viral life cycle, but not the earlier steps such as reverse transcription or integration. Immunoprecipitation assays revealed that ABIN1 specifically inhibits the proto-oncogene HDM2 catalyzed K63-linked polyubiquitination of Tat at Lys71, which is critical for the transactivation activity of Tat. The ubiquitin chain binding activity of ABIN1 carried by UBAN domain turned out to be essential for the inhibitory role of ABIN1. The results of immunofluorescence localization experiments suggested that ABIN1 may obstruct Tat ubiquitination by redistributing some of HDM2 from the nucleus to the cytoplasm.
Conclusions: Our findings have revealed ABIN1 as intrinsic suppressor of HIV-1 mRNA transcription by regulating the ubiquitination of Tat.
Keywords: ABIN1; HDM2; HIV-1; Tat; Transcription; Ubiquitination.
Figures
Similar articles
-
FBXO45 restricts HIV-1 replication by inducing SQSTM1/p62-mediated autophagic degradation of Tat.J Virol. 2025 Mar 18;99(3):e0191224. doi: 10.1128/jvi.01912-24. Epub 2025 Feb 12. J Virol. 2025. PMID: 39936917 Free PMC article.
-
HIV-1 Tat Recruits HDM2 E3 Ligase To Target IRF-1 for Ubiquitination and Proteasomal Degradation.mBio. 2016 Oct 18;7(5):e01528-16. doi: 10.1128/mBio.01528-16. mBio. 2016. PMID: 27795392 Free PMC article.
-
Specific Inhibition of HIV Infection by the Action of Spironolactone in T Cells.J Virol. 2016 Nov 14;90(23):10972-10980. doi: 10.1128/JVI.01722-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27681137 Free PMC article.
-
The role of Tat in HIV-1 replication: an activator and/or a suppressor?AIDS Rev. 2002 Jan-Mar;4(1):41-9. AIDS Rev. 2002. PMID: 11998784 Review.
-
Tat basic domain: A "Swiss army knife" of HIV-1 Tat?Rev Med Virol. 2019 Mar;29(2):e2031. doi: 10.1002/rmv.2031. Epub 2019 Jan 4. Rev Med Virol. 2019. PMID: 30609200 Review.
Cited by
-
Deubiquitinating Enzyme USP21 Inhibits HIV-1 Replication by Downregulating Tat Expression.J Virol. 2021 Jun 10;95(13):e0046021. doi: 10.1128/JVI.00460-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33827943 Free PMC article.
-
FBXO45 restricts HIV-1 replication by inducing SQSTM1/p62-mediated autophagic degradation of Tat.J Virol. 2025 Mar 18;99(3):e0191224. doi: 10.1128/jvi.01912-24. Epub 2025 Feb 12. J Virol. 2025. PMID: 39936917 Free PMC article.
-
Neuroinflammation generated by HIV-infected microglia promotes dysfunction and death of neurons in human brain organoids.PNAS Nexus. 2024 Apr 29;3(5):pgae179. doi: 10.1093/pnasnexus/pgae179. eCollection 2024 May. PNAS Nexus. 2024. PMID: 38737767 Free PMC article.
-
Ubiquitination and SUMOylation in HIV Infection: Friends and Foes.Curr Issues Mol Biol. 2020;35:159-194. doi: 10.21775/cimb.035.159. Epub 2019 Aug 18. Curr Issues Mol Biol. 2020. PMID: 31422939 Free PMC article. Review.
-
Effect of transcription inhibition and generation of suppressive viral non-coding RNAs.Retrovirology. 2019 Apr 29;16(1):13. doi: 10.1186/s12977-019-0475-0. Retrovirology. 2019. PMID: 31036006 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

