Transcriptional Mechanisms of Resistance to Anti-PD-1 Therapy
- PMID: 28193624
- PMCID: PMC5474192
- DOI: 10.1158/1078-0432.CCR-17-0270
Transcriptional Mechanisms of Resistance to Anti-PD-1 Therapy
Abstract
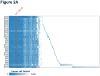
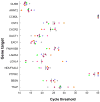
Purpose: To explore factors associated with response and resistance to anti-PD-1 therapy, we analyzed multiple disease sites at autopsy in a patient with widely metastatic melanoma who had a heterogeneous response.Materials and Methods: Twenty-six melanoma specimens (four premortem, 22 postmortem) were subjected to whole exome sequencing. Candidate immunologic markers and gene expression were assessed in 10 cutaneous metastases showing response or progression during therapy.Results: The melanoma was driven by biallelic inactivation of NF1 All lesions had highly concordant mutational profiles and copy number alterations, indicating linear clonal evolution. Expression of candidate immunologic markers was similar in responding and progressing lesions. However, progressing cutaneous metastases were associated with overexpression of genes associated with extracellular matrix and neutrophil function.Conclusions: Although mutational and immunologic differences have been proposed as the primary determinants of heterogeneous response/resistance to targeted therapies and immunotherapies, respectively, differential lesional gene expression profiles may also dictate anti-PD-1 outcomes. Clin Cancer Res; 23(12); 3168-80. ©2017 AACRSee related commentary by Wilmott et al., p. 2921.
©2017 American Association for Cancer Research.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

