Contribution of epigenetic mechanisms to variation in cancer risk among tissues
- PMID: 28193856
- PMCID: PMC5338490
- DOI: 10.1073/pnas.1616556114
Contribution of epigenetic mechanisms to variation in cancer risk among tissues
Abstract
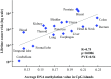
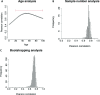
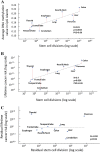
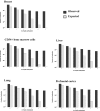
Recently, it was suggested that tissue variation in cancer risk originates from differences in the number of stem-cell divisions underlying each tissue, leading to different mutation loads. We show that this variation is also correlated with the degree of aberrant CpG island DNA methylation in normal cells. Methylation accumulates during aging in a subset of molecules, suggesting that the epigenetic landscape within a founder-cell population may contribute to tumor formation.
Keywords: aging; methylation; polycomb.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M. Comparative Risk Assessment collaborating group (Cancers) Causes of cancer in the world: Comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366(9499):1784–1793. - PubMed
-
- Fearon ER. Human cancer syndromes: Clues to the origin and nature of cancer. Science. 1997;278(5340):1043–1050. - PubMed
-
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

