Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding
- PMID: 28193899
- PMCID: PMC5338521
- DOI: 10.1073/pnas.1612927114
Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding
Abstract
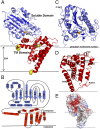
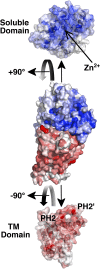
Multidrug-resistant (MDR) gram-negative bacteria have increased the prevalence of fatal sepsis in modern times. Colistin is a cationic antimicrobial peptide (CAMP) antibiotic that permeabilizes the bacterial outer membrane (OM) and has been used to treat these infections. The OM outer leaflet is comprised of endotoxin containing lipid A, which can be modified to increase resistance to CAMPs and prevent clearance by the innate immune response. One type of lipid A modification involves the addition of phosphoethanolamine to the 1 and 4' headgroup positions by phosphoethanolamine transferases. Previous structural work on a truncated form of this enzyme suggested that the full-length protein was required for correct lipid substrate binding and catalysis. We now report the crystal structure of a full-length lipid A phosphoethanolamine transferase from Neisseria meningitidis, determined to 2.75-Å resolution. The structure reveals a previously uncharacterized helical membrane domain and a periplasmic facing soluble domain. The domains are linked by a helix that runs along the membrane surface interacting with the phospholipid head groups. Two helices located in a periplasmic loop between two transmembrane helices contain conserved charged residues and are implicated in substrate binding. Intrinsic fluorescence, limited proteolysis, and molecular dynamics studies suggest the protein may sample different conformational states to enable the binding of two very different- sized lipid substrates. These results provide insights into the mechanism of endotoxin modification and will aid a structure-guided rational drug design approach to treating multidrug-resistant bacterial infections.
Keywords: Neisseria; lipid modification; membrane protein structure; molecular dynamics; multidrug resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
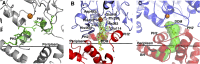
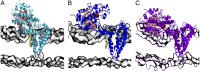
References
-
- World Health Organization . Antimicrobial resistance: Global report on surveillance. WHO; Geneva: 2014. pp. 1–256.
-
- US Department of Health and Human Services . Antibiotic resistance threats in the United States 2013. Centers for Disease Control and Prevention; Atlanta: 2013. pp. 1–113.
-
- Osei Sekyere J, Govinden U, Bester LA, Essack SY. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J Appl Microbiol. 2016;121(3):601–617. - PubMed
-
- Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

