Structural Analysis of Mycobacterium tuberculosis Homologues of the Eukaryotic Proteasome Assembly Chaperone 2 (PAC2)
- PMID: 28193903
- PMCID: PMC5388811
- DOI: 10.1128/JB.00846-16
Structural Analysis of Mycobacterium tuberculosis Homologues of the Eukaryotic Proteasome Assembly Chaperone 2 (PAC2)
Abstract
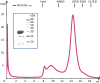
A previous bioinformatics analysis identified the Mycobacterium tuberculosis proteins Rv2125 and Rv2714 as orthologs of the eukaryotic proteasome assembly chaperone 2 (PAC2). We set out to investigate whether Rv2125 or Rv2714 can function in proteasome assembly. We solved the crystal structure of Rv2125 at a resolution of 3.0 Å, which showed an overall fold similar to that of the PAC2 family proteins that include the archaeal PbaB and the yeast Pba1. However, Rv2125 and Rv2714 formed trimers, whereas PbaB forms tetramers and Pba1 dimerizes with Pba2. We also found that purified Rv2125 and Rv2714 could not bind to M. tuberculosis 20S core particles. Finally, proteomic analysis showed that the levels of known proteasome components and substrate proteins were not affected by disruption of Rv2125 in M. tuberculosis Our work suggests that Rv2125 does not participate in bacterial proteasome assembly or function.IMPORTANCE Although many bacteria do not encode proteasomes, M. tuberculosis not only uses proteasomes but also has evolved a posttranslational modification system called pupylation to deliver proteins to the proteasome. Proteasomes are essential for M. tuberculosis to cause lethal infections in animals; thus, determining how proteasomes are assembled may help identify new ways to combat tuberculosis. We solved the structure of a predicted proteasome assembly factor, Rv2125, and isolated a genetic Rv2125 mutant of M. tuberculosis Our structural, biochemical, and genetic studies indicate that Rv2125 and Rv2714 do not function as proteasome assembly chaperones and are unlikely to have roles in proteasome biology in mycobacteria.
Keywords: Mycobacterium tuberculosis; proteasome; protein chaperone; structural biology.
Copyright © 2017 American Society for Microbiology.
Figures
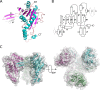

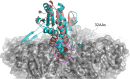

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

