MOBAS: identification of disease-associated protein subnetworks using modularity-based scoring
- PMID: 28194175
- PMCID: PMC5270451
- DOI: 10.1186/s13637-015-0025-6
MOBAS: identification of disease-associated protein subnetworks using modularity-based scoring
Abstract
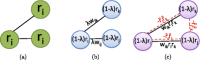
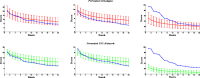
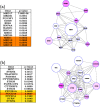
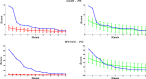
Network-based analyses are commonly used as powerful tools to interpret the findings of genome-wide association studies (GWAS) in a functional context. In particular, identification of disease-associated functional modules, i.e., highly connected protein-protein interaction (PPI) subnetworks with high aggregate disease association, are shown to be promising in uncovering the functional relationships among genes and proteins associated with diseases. An important issue in this regard is the scoring of subnetworks by integrating two quantities: disease association of individual gene products and network connectivity among proteins. Current scoring schemes either disregard the level of connectivity and focus on the aggregate disease association of connected proteins or use a linear combination of these two quantities. However, such scoring schemes may produce arbitrarily large subnetworks which are often not statistically significant or require tuning of parameters that are used to weigh the contributions of network connectivity and disease association. Here, we propose a parameter-free scoring scheme that aims to score subnetworks by assessing the disease association of interactions between pairs of gene products. We also incorporate the statistical significance of network connectivity and disease association into the scoring function. We test the proposed scoring scheme on a GWAS dataset for two complex diseases type II diabetes (T2D) and psoriasis (PS). Our results suggest that subnetworks identified by commonly used methods may fail tests of statistical significance after correction for multiple hypothesis testing. In contrast, the proposed scoring scheme yields highly significant subnetworks, which contain biologically relevant proteins that cannot be identified by analysis of genome-wide association data alone. We also show that the proposed scoring scheme identifies subnetworks that are reproducible across different cohorts, and it can robustly recover relevant subnetworks at lower sampling rates.
Keywords: Genome-wide association studies; Protein-protein interaction network; Statistical significance.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

