Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential
- PMID: 28195173
- PMCID: PMC5316866
- DOI: 10.1038/ncomms14428
Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential
Abstract
The recent identification of progenitor populations that contribute to the developing heart in a distinct spatial and temporal manner has fundamentally improved our understanding of cardiac development. However, the mechanisms that direct atrial versus ventricular specification remain largely unknown. Here we report the identification of a progenitor population that gives rise primarily to cardiovascular cells of the ventricles and only to few atrial cells (<5%) of the differentiated heart. These progenitors are specified during gastrulation, when they transiently express Foxa2, a gene not previously implicated in cardiac development. Importantly, Foxa2+ cells contribute to previously identified progenitor populations in a defined pattern and ratio. Lastly, we describe an analogous Foxa2+ population during differentiation of embryonic stem cells. Together, these findings provide insight into the developmental origin of ventricular and atrial cells, and may lead to the establishment of new strategies for generating chamber-specific cell types from pluripotent stem cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




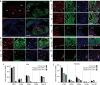
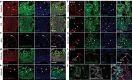



References
-
- Buckingham M., Meilhac S. & Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826–835 (2005). - PubMed
-
- Arias E., Kochanek K. D., Xu J. & Murphy S. L. Mortality in the United States, 2014. NCHS Data Brief 229, 1–8 (2015). - PubMed
-
- Bruneau B. G. & Srivastava D. Congenital heart disease: entering a new era of human genetics. Circ. Res. 114, 598–599 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

