Activity-based probes for the ubiquitin conjugation-deconjugation machinery: new chemistries, new tools, and new insights
- PMID: 28196299
- PMCID: PMC7163952
- DOI: 10.1111/febs.14039
Activity-based probes for the ubiquitin conjugation-deconjugation machinery: new chemistries, new tools, and new insights
Abstract
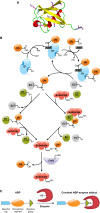
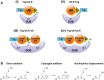
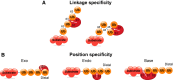
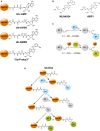
The reversible post-translational modification of proteins by ubiquitin and ubiquitin-like proteins regulates almost all cellular processes, by affecting protein degradation, localization, and complex formation. Deubiquitinases (DUBs) are proteases that remove ubiquitin modifications or cleave ubiquitin chains. Most DUBs are cysteine proteases, which makes them well suited for study by activity-based probes. These DUB probes report on deubiquitinase activity by reacting covalently with the active site in an enzyme-catalyzed manner. They have proven to be important tools to study DUB selectivity and proteolytic activity in different settings, to identify novel DUBs, and to characterize deubiquitinase inhibitors. Inspired by the efficacy of activity-based probes for DUBs, several groups have recently reported probes for the ubiquitin conjugation machinery (E1, E2, and E3 enzymes). Many of these enzymes, while not proteases, also posses active site cysteine residues and can be targeted by covalent probes. In this review, we will discuss how features of the probe (cysteine-reactive group, recognition element, and reporter tag) affect reactivity and suitability for certain experimental applications. We will also review the diverse applications of the current probes, and discuss the need for new probe types to study emerging aspects of ubiquitin biology.
Keywords: E1; E3; activity-based probe; activity-based protein profiling; chemoproteomics; deubiquitinase; protease; ubiquitin; ubiquitin-proteasome system.
© 2017 Federation of European Biochemical Societies.
Figures
References
-
- Hershko A & Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67, 425–479. - PubMed
-
- Komander D (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37, 937–953. - PubMed
-
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D & Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21, 921–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

