Leveraging Online Resources to Prioritize Candidate Genes for Functional Analyses: Using the Fetal Testis as a Test Case
- PMID: 28196369
- PMCID: PMC6171109
- DOI: 10.1159/000455113
Leveraging Online Resources to Prioritize Candidate Genes for Functional Analyses: Using the Fetal Testis as a Test Case
Abstract
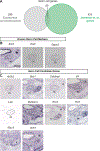
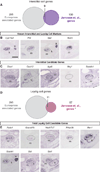
With each new microarray or RNA-seq experiment, massive quantities of transcriptomic information are generated with the purpose to produce a list of candidate genes for functional analyses. Yet an effective strategy remains elusive to prioritize the genes on these candidate lists. In this review, we outline a prioritizing strategy by taking a step back from the bench and leveraging the rich range of public databases. This in silico approach provides an economical, less biased, and more effective solution. We discuss the publicly available online resources that can be used to answer a range of questions about a gene. Is the gene of interest expressed in the system of interest (using expression databases)? Where else is this gene expressed (using added-value transcriptomic resources)? What pathways and processes is the gene involved in (using enriched gene pathway analysis and mouse knockout databases)? Is this gene correlated with human diseases (using human disease variant databases)? Using mouse fetal testis as an example, our strategies identified 298 genes annotated as expressed in the fetal testis. We cross-referenced these genes to existing microarray data and narrowed the list down to cell-type-specific candidates (35 for Sertoli cells, 11 for Leydig cells, and 25 for germ cells). Our strategies can be customized so that they allow researchers to effectively and confidently prioritize genes for functional analysis.
© 2017 S. Karger AG, Basel.
Figures


References
-
- Ono M, Harley VR: Disorders of sex development: new genes, new concepts. Nat Rev Endocrinol 9:79–91 (2013). - PubMed
-
- McClelland KS, Bell K, Larney C, Harley VR, Sinclair AH, Oshlack A, et al.: Purification and Transcriptomic Analysis of Mouse Fetal Leydig Cells Reveals Candidate Genes for Specification of Gonadal Steroidogenic Cells. Biol Reprod 92:145–145 (2015). - PubMed
-
- Beverdam A, Koopman P: Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet 15:417–431 (2006). - PubMed
-
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, et al.: Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol 287:361–377 (2005). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

