Alu-Derived Alternative Splicing Events Specific to Macaca Lineages in CTSF Gene
- PMID: 28196413
- PMCID: PMC5339500
- DOI: 10.14348/molcells.2017.2204
Alu-Derived Alternative Splicing Events Specific to Macaca Lineages in CTSF Gene
Abstract
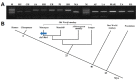
Cathepsin F, which is encoded by CTSF, is a cysteine proteinase ubiquitously expressed in several tissues. In a previous study, novel transcripts of the CTSF gene were identified in the crab-eating monkey deriving from the integration of an Alu element-AluYRa1. The occurrence of AluYRa1-derived alternative transcripts and the mechanism of exonization events in the CTSF gene of human, rhesus monkey, and crab-eating monkey were investigated using PCR and reverse transcription PCR on the genomic DNA and cDNA isolated from several tissues. Results demonstrated that AluYRa1 was only integrated into the genome of Macaca species and this lineage-specific integration led to exonization events by producing a conserved 3' splice site. Six transcript variants (V1-V6) were generated by alternative splicing (AS) events, including intron retention and alternative 5' splice sites in the 5' and 3' flanking regions of CTSF_AluYRa1. Among them, V3-V5 transcripts were ubiquitously expressed in all tissues of rhesus monkey and crab-eating monkey, whereas AluYRa1-exonized V1 was dominantly expressed in the testis of the crab-eating monkey, and V2 was only expressed in the testis of the two monkeys. These five transcript variants also had different amino acid sequences in the C-terminal region of CTSF, as compared to reference sequences. Thus, species-specific Alu-derived exonization by lineage-specific integration of Alu elements and AS events seems to have played an important role during primate evolution by producing transcript variants and gene diversification.
Keywords: Alternative splicing; Alu; CTSF; exonization; primate.
Figures





References
-
- Ahn S.J., Kim N.Y., Seo J.S., Je J.E., Sung J.H., Lee S.H., Kim M.S., Kim J.K., Chung J.K., Lee H.H. Molecular cloning, mRNA expression and enzymatic characterization of cathepsin F from olive flounder (Paralichthys olivaceus) Comp Biochem Physiol B Biochem Mol Biol. 2009;154:211–220. - PubMed
-
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. - PubMed
-
- Carlsson H.E., Schapiro S.J., Farah I., Hau J. Use of primates in research: a global overview. Am J Primatol. 2004;63:225–237. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

