2.7 million samples genotyped for HLA by next generation sequencing: lessons learned
- PMID: 28196473
- PMCID: PMC5309984
- DOI: 10.1186/s12864-017-3575-z
2.7 million samples genotyped for HLA by next generation sequencing: lessons learned
Abstract
Background: At the DKMS Life Science Lab, Next Generation Sequencing (NGS) has been used for ultra-high-volume high-resolution genotyping of HLA loci for the last three and a half years. Here, we report on our experiences in genotyping the HLA, CCR5, ABO, RHD and KIR genes using a direct amplicon sequencing approach on Illumina MiSeq and HiSeq 2500 instruments.
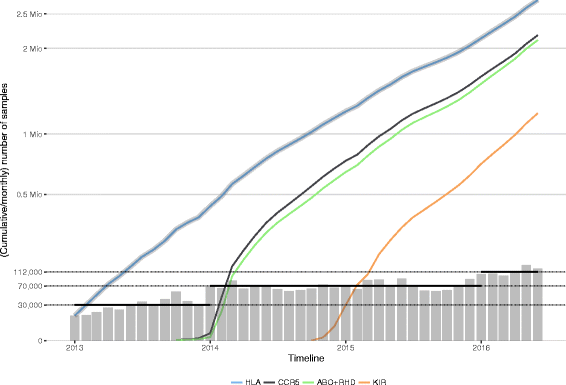
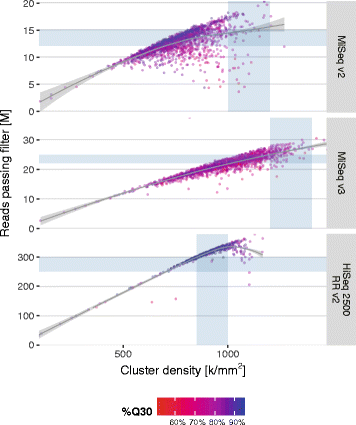
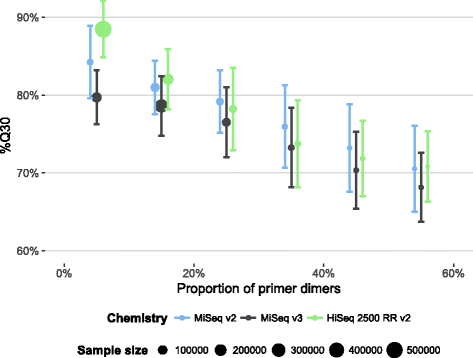
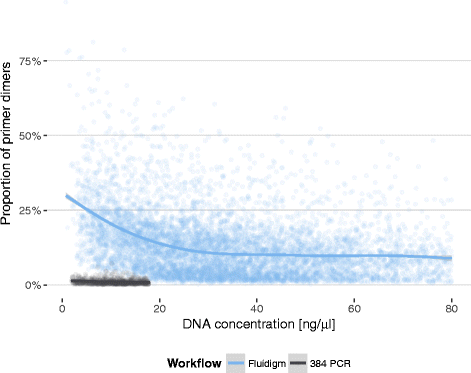
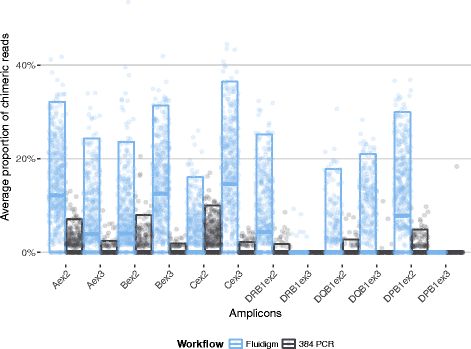
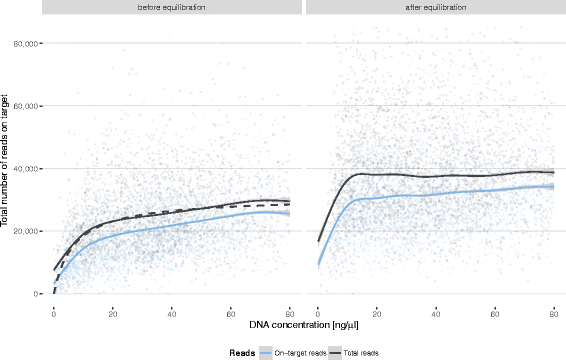
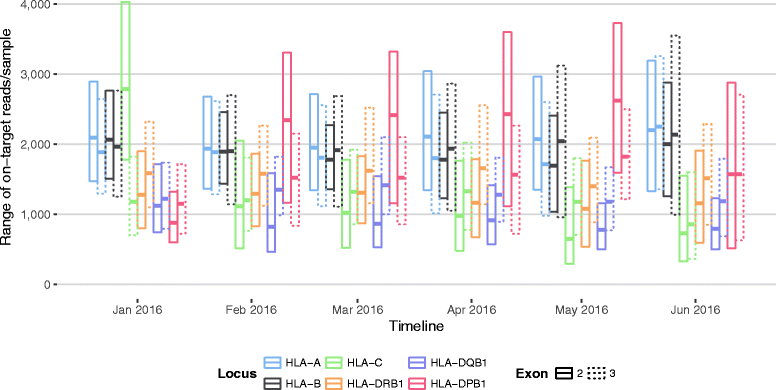
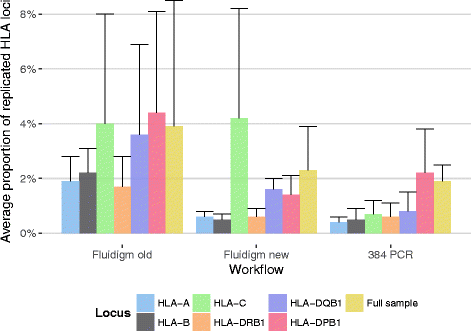
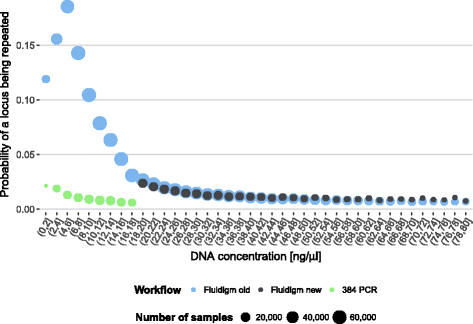
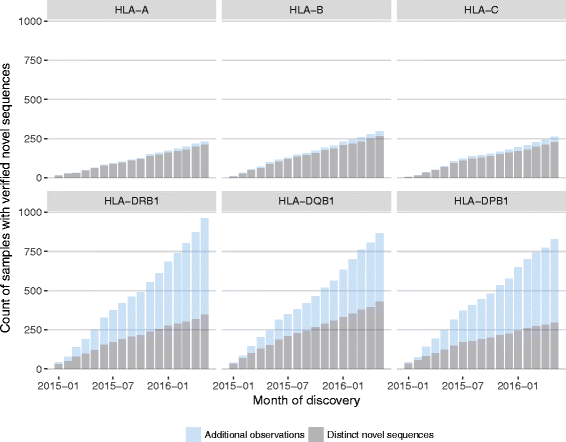
Results: Between January 2013 and June 2016, 2,714,110 samples largely from German, Polish and UK-based potential stem cell donors have been processed. 98.9% of all alleles for the targeted HLA loci (HLA-A, -B, -C, -DRB1, -DQB1 and -DPB1) were typed at high resolution or better. Initially a simple three-step workflow based on nanofluidic chips in conjunction with 4-primer amplicon tagging was used. Over time, we found that this setup results in PCR artefacts such as primer dimers and PCR-mediated recombination, which may necessitate repeat typing. Split workflows for low- and high-DNA-concentration samples helped alleviate these problems and reduced average per-locus repeat rates from 3.1 to 1.3%. Further optimisations of the workflow included the use of phosphorothioate oligos to reduce primer degradation and primer dimer formation, and employing statistical models to predict read yield from initial template DNA concentration to avoid intermediate quantification of PCR products. Finally, despite the populations typed at DKMS Life Science Lab being relatively homogenous genetically, an analysis of 1.4 million donors processed between January 2015 and May 2016 led to the discovery of 1,919 distinct novel HLA alleles.
Conclusions: Amplicon-based NGS HLA genotyping workflows have become the workhorse in high-volume tissue typing of registry donors. The optimisation of workflow practices over multiple years has led to insights and solutions that improve the efficiency and robustness of short amplicon based genotyping workflows.
Keywords: Amplicon PCR; DKMS; HLA genotyping; High resolution; High throughput; Next generation sequencing; Novel alleles; PCR chimerism; Primer dimers.
Figures

References
-
- Loiseau P, Busson M, Balere M-L, Dormoy A, Bignon J-D, Gagne K, et al. HLA association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant. 2007;13:965–74. doi: 10.1016/j.bbmt.2007.04.010. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

