Using Quantitative Spectrometry to Understand the Influence of Genetics and Nutritional Perturbations On the Virulence Potential of Staphylococcus aureus
- PMID: 28196877
- PMCID: PMC5393389
- DOI: 10.1074/mcp.O116.065581
Using Quantitative Spectrometry to Understand the Influence of Genetics and Nutritional Perturbations On the Virulence Potential of Staphylococcus aureus
Abstract
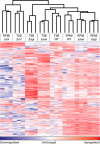
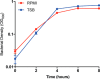
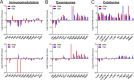
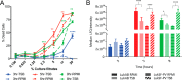
Staphylococcus aureus (Sa) is the leading cause of a variety of bacterial infections ranging from superficial skin infections to invasive and life threatening diseases such as septic bacteremia, necrotizing pneumonia, and endocarditis. The success of Sa as a human pathogen is contributed to its ability to adapt to different environments by changing expression, production, or secretion of virulence factors. Although Sa immune evasion is well-studied, the regulation of virulence factors under different nutrient and growth conditions is still not well understood. Here, we used label-free quantitative mass spectrometry to quantify and compare the Sa exoproteins (i.e. exoproteomes) of master regulator mutants or established reference strains. Different environmental conditions were addressed by growing the bacteria in rich or minimal media at different phases of growth. We observed clear differences in the composition of the exoproteomes depending on the genetic background or growth conditions. The relative abundance of cytotoxins determined in our study correlated well with differences in cytotoxicity measured by lysis of human neutrophils. Our findings demonstrate that label-free quantitative mass spectrometry is a versatile tool for predicting the virulence of bacterial strains and highlights the importance of the experimental design for in vitro studies. Furthermore, the results indicate that label-free proteomics can be used to cluster isolates into groups with similar virulence properties, highlighting the power of label-free quantitative mass spectrometry to distinguish Sa strains.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Diekema D. J., Pfaller M. A., Schmitz F. J., Smayevsky J., Bell J., Jones R. N., Beach M., and Group S. P. (2001) Survey of infections because of Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 32, S114–S132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

