Redundant Postsynaptic Functions of SynCAMs 1-3 during Synapse Formation
- PMID: 28197078
- PMCID: PMC5281628
- DOI: 10.3389/fnmol.2017.00024
Redundant Postsynaptic Functions of SynCAMs 1-3 during Synapse Formation
Abstract
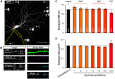
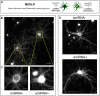
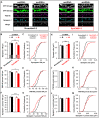
Investigating the roles of synaptogenic adhesion molecules during synapse formation has proven challenging, often due to compensatory functions between additional family members. The synaptic cell adhesion molecules 1-3 (SynCAM1-3) are expressed both pre- and postsynaptically, share highly homologous domains and are synaptogenic when ectopically presented to neurons; yet their endogenous functions during synaptogenesis are unclear. Here we report that SynCAM1-3 are functionally redundant and collectively necessary for synapse formation in cultured hippocampal neurons. Only triple knockdown (KD) of SynCAM1-3 using highly efficient, chained artificial microRNAs (amiRNAs) reduced synapse density and increased synapse area. Electrophysiological recordings of quantal release events supported an increase in synapse size caused by SynCAM1-3 depletion. Furthermore, a combinatorial, mosaic lentiviral approach comparing wild type (WT) and SynCAM1-3 KD neurons in the same culture demonstrate that SynCAM1-3 set synapse number and size through postsynaptic mechanisms. The results demonstrate that the redundancy between SynCAM1-3 has concealed their synaptogenic function at the postsynaptic terminal.
Keywords: SynCAM; adhesion; artificial miRNA; hippocampus; knockdown; mosaic; redundancy; synapse formation.
Figures


Similar articles
-
Identification of Protein Tyrosine Phosphatase Receptor Type O (PTPRO) as a Synaptic Adhesion Molecule that Promotes Synapse Formation.J Neurosci. 2017 Oct 11;37(41):9828-9843. doi: 10.1523/JNEUROSCI.0729-17.2017. Epub 2017 Sep 4. J Neurosci. 2017. PMID: 28871037 Free PMC article.
-
Establishment of transgenic fluorescent mice for labeling synapses and screening synaptogenic adhesion molecules.Elife. 2024 Mar 7;13:e81884. doi: 10.7554/eLife.81884. Elife. 2024. PMID: 38450720 Free PMC article.
-
Down-regulation of mRNAs for synaptic adhesion molecules neuroligin-2 and -3 and synCAM1 in spinal motoneurons after axotomy.J Comp Neurol. 2007 Jul 10;503(2):308-18. doi: 10.1002/cne.21382. J Comp Neurol. 2007. PMID: 17492651
-
SynCAMs extend their functions beyond the synapse.Eur J Neurosci. 2014 Jun;39(11):1752-60. doi: 10.1111/ejn.12544. Epub 2014 Mar 15. Eur J Neurosci. 2014. PMID: 24628990 Review.
-
SynCAMs - From axon guidance to neurodevelopmental disorders.Mol Cell Neurosci. 2017 Jun;81:41-48. doi: 10.1016/j.mcn.2016.08.012. Epub 2016 Sep 1. Mol Cell Neurosci. 2017. PMID: 27594578 Review.
Cited by
-
Mathematical model of mechanobiology of acute and repeated synaptic injury and systemic biomarker kinetics.Front Cell Neurosci. 2023 Feb 6;17:1007062. doi: 10.3389/fncel.2023.1007062. eCollection 2023. Front Cell Neurosci. 2023. PMID: 36814869 Free PMC article.
-
Axoglial Adhesion by Cadm4 Regulates CNS Myelination.Neuron. 2019 Jan 16;101(2):224-231.e5. doi: 10.1016/j.neuron.2018.11.032. Epub 2018 Dec 11. Neuron. 2019. PMID: 30551998 Free PMC article.
-
4.1Ba is necessary for glutamatergic synapse formation in the sensorimotor circuit of developing zebrafish.PLoS One. 2018 Oct 4;13(10):e0205255. doi: 10.1371/journal.pone.0205255. eCollection 2018. PLoS One. 2018. PMID: 30286167 Free PMC article.
-
Oligodendrocytes express synaptic proteins that modulate myelin sheath formation.Nat Commun. 2019 Sep 11;10(1):4125. doi: 10.1038/s41467-019-12059-y. Nat Commun. 2019. PMID: 31511515 Free PMC article.
-
Differential Contribution of Cadm1-Cadm3 Cell Adhesion Molecules to Peripheral Myelinated Axons.J Neurosci. 2021 Feb 17;41(7):1393-1400. doi: 10.1523/JNEUROSCI.2736-20.2020. Epub 2021 Jan 4. J Neurosci. 2021. PMID: 33397712 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

