Characterization of the Genomic Diversity of Norovirus in Linked Patients Using a Metagenomic Deep Sequencing Approach
- PMID: 28197136
- PMCID: PMC5282449
- DOI: 10.3389/fmicb.2017.00073
Characterization of the Genomic Diversity of Norovirus in Linked Patients Using a Metagenomic Deep Sequencing Approach
Abstract
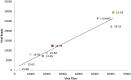
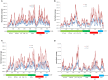
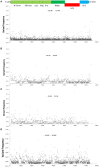
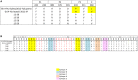
Norovirus (NoV) is the leading cause of gastroenteritis worldwide. A robust cell culture system does not exist for NoV and therefore detailed characterization of outbreak and sporadic strains relies on molecular techniques. In this study, we employed a metagenomic approach that uses non-specific amplification followed by next-generation sequencing to whole genome sequence NoV genomes directly from clinical samples obtained from 8 linked patients. Enough sequencing depth was obtained for each sample to use a de novo assembly of near-complete genome sequences. The resultant consensus sequences were then used to identify inter-host nucleotide variations that occur after direct transmission, analyze amino acid variations in the major capsid protein, and provide evidence of recombination events. The analysis of intra-host quasispecies diversity was possible due to high coverage-depth. We also observed a linear relationship between NoV viral load in the clinical sample and the number of sequence reads that could be attributed to NoV. The method demonstrated here has the potential for future use in whole genome sequence analyses of other RNA viruses isolated from clinical, environmental, and food specimens.
Keywords: antigenic variation; genetic variation; linked patients; next-generation sequencing (NGS); norovirus; recombination.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

