Isolation, Characterization and Transcriptome Analysis of a Cytokinin Receptor Mutant Osckt1 in Rice
- PMID: 28197164
- PMCID: PMC5281565
- DOI: 10.3389/fpls.2017.00088
Isolation, Characterization and Transcriptome Analysis of a Cytokinin Receptor Mutant Osckt1 in Rice
Abstract
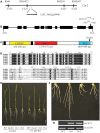
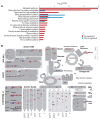
Cytokinins play important roles in regulating plant development, including shoot and root meristems, leaf longevity, and grain yield. However, the in planta functions of rice cytokinin receptors have not been genetically characterized yet. Here we isolated a rice mutant, Osckt1, with enhanced tolerance to cytokinin treatment. Further analysis showed that Osckt1 was insensitive to aromatic cytokinins but responded normally to isoprenoid and phenylurea-type cytokinins. Map-based cloning revealed that the mutation occurred in a putative cytokinin receptor gene, histidine kinase 6 (OsHK6). OsCKT1 was found to be expressed in various tissues throughout the plant and the protein was located in the endoplasmic reticulum. In addition, whole-genome gene expression profiling analysis showed that OsCKT1 was involved in cytokinin regulation of a number of biological processes, including secondary metabolism, sucrose and starch metabolism, chlorophyll synthesis, and photosynthesis. Our results demonstrate that OsCKT1 plays important roles in cytokinin perception and control of root development in rice.
Keywords: Oryza sativa; cytokinin; histidine kinase; rice; transcriptome.
Figures



Similar articles
-
Functional identification of OsHk6 as a homotypic cytokinin receptor in rice with preferential affinity for iP.Plant Cell Physiol. 2012 Jul;53(7):1334-43. doi: 10.1093/pcp/pcs079. Epub 2012 May 28. Plant Cell Physiol. 2012. PMID: 22642989
-
ABC transporter OsABCG18 controls the shootward transport of cytokinins and grain yield in rice.J Exp Bot. 2019 Nov 18;70(21):6277-6291. doi: 10.1093/jxb/erz382. J Exp Bot. 2019. PMID: 31504730 Free PMC article.
-
A semi-dominant mutation in the gene encoding histidine kinase influences rice morphology.Genes Genet Syst. 2025 Apr 4;100. doi: 10.1266/ggs.24-00223. Epub 2025 Feb 6. Genes Genet Syst. 2025. PMID: 39909425
-
The diverse roles of cytokinins in regulating leaf development.Hortic Res. 2021 Jun 1;8(1):118. doi: 10.1038/s41438-021-00558-3. Hortic Res. 2021. PMID: 34059666 Free PMC article. Review.
-
Cytokinin dehydrogenase: a genetic target for yield improvement in wheat.Plant Biotechnol J. 2020 Mar;18(3):614-630. doi: 10.1111/pbi.13305. Epub 2019 Dec 22. Plant Biotechnol J. 2020. PMID: 31782596 Free PMC article. Review.
Cited by
-
The cytokinin receptor OHK4/OsHK4 regulates inflorescence architecture in rice via an IDEAL PLANT ARCHITECTURE1/WEALTHY FARMER'S PANICLE-mediated positive feedback circuit.Plant Cell. 2023 Dec 21;36(1):40-64. doi: 10.1093/plcell/koad257. Plant Cell. 2023. PMID: 37811656 Free PMC article.
-
Understanding plant-microbe interaction of rice and soybean with two contrasting diazotrophic bacteria through comparative transcriptome analysis.Front Plant Sci. 2022 Nov 18;13:939395. doi: 10.3389/fpls.2022.939395. eCollection 2022. Front Plant Sci. 2022. PMID: 36483966 Free PMC article.
-
Uncovering the Genomic Regions Associated with Yield Maintenance in Rice Under Drought Stress Using an Integrated Meta-Analysis Approach.Rice (N Y). 2024 Jan 16;17(1):7. doi: 10.1186/s12284-024-00684-1. Rice (N Y). 2024. PMID: 38227151 Free PMC article.
-
Understanding the Intricate Web of Phytohormone Signalling in Modulating Root System Architecture.Int J Mol Sci. 2021 May 24;22(11):5508. doi: 10.3390/ijms22115508. Int J Mol Sci. 2021. PMID: 34073675 Free PMC article. Review.
-
Endoplasmic Reticulum-Localized PURINE PERMEASE1 Regulates Plant Height and Grain Weight by Modulating Cytokinin Distribution in Rice.Front Plant Sci. 2020 Dec 22;11:618560. doi: 10.3389/fpls.2020.618560. eCollection 2020. Front Plant Sci. 2020. PMID: 33414802 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

