Using Global Analysis to Extend the Accuracy and Precision of Binding Measurements with T cell Receptors and Their Peptide/MHC Ligands
- PMID: 28197404
- PMCID: PMC5281623
- DOI: 10.3389/fmolb.2017.00002
Using Global Analysis to Extend the Accuracy and Precision of Binding Measurements with T cell Receptors and Their Peptide/MHC Ligands
Abstract
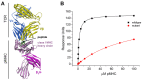
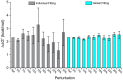
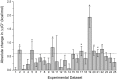
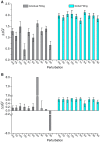
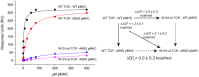
In cellular immunity, clonally distributed T cell receptors (TCRs) engage complexes of peptides bound to major histocompatibility complex proteins (pMHCs). In the interactions of TCRs with pMHCs, regions of restricted and variable diversity align in a structurally complex fashion. Many studies have used mutagenesis to attempt to understand the "roles" played by various interface components in determining TCR recognition properties such as specificity and cross-reactivity. However, these measurements are often complicated or even compromised by the weak affinities TCRs maintain toward pMHC. Here, we demonstrate how global analysis of multiple datasets can be used to significantly extend the accuracy and precision of such TCR binding experiments. Application of this approach should positively impact efforts to understand TCR recognition and facilitate the creation of mutational databases to help engineer TCRs with tuned molecular recognition properties. We also show how global analysis can be used to analyze double mutant cycles in TCR-pMHC interfaces, which can lead to new insights into immune recognition.
Keywords: T cell receptor; binding; double mutant cycle; global analysis; mutagenesis; peptide/MHC.
Figures

Similar articles
-
Conserved Vδ1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by δ/αβ T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.J Virol. 2017 Aug 10;91(17):e00725-17. doi: 10.1128/JVI.00725-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615212 Free PMC article.
-
The specificity of TCR/pMHC interaction.Curr Opin Immunol. 2002 Feb;14(1):52-65. doi: 10.1016/s0952-7915(01)00298-9. Curr Opin Immunol. 2002. PMID: 11790533 Review.
-
A study of CDR3 loop dynamics reveals distinct mechanisms of peptide recognition by T-cell receptors exhibiting different levels of cross-reactivity.Immunology. 2018 Apr;153(4):466-478. doi: 10.1111/imm.12849. Epub 2017 Nov 8. Immunology. 2018. PMID: 28992359 Free PMC article.
-
Stress-testing the relationship between T cell receptor/peptide-MHC affinity and cross-reactivity using peptide velcro.Proc Natl Acad Sci U S A. 2018 Jul 31;115(31):E7369-E7378. doi: 10.1073/pnas.1802746115. Epub 2018 Jul 18. Proc Natl Acad Sci U S A. 2018. PMID: 30021852 Free PMC article.
-
Structure-Based, Rational Design of T Cell Receptors.Front Immunol. 2013 Sep 12;4:268. doi: 10.3389/fimmu.2013.00268. Front Immunol. 2013. PMID: 24062738 Free PMC article. Review.
Cited by
-
An Engineered T Cell Receptor Variant Realizes the Limits of Functional Binding Modes.Biochemistry. 2020 Nov 3;59(43):4163-4175. doi: 10.1021/acs.biochem.0c00689. Epub 2020 Oct 19. Biochemistry. 2020. PMID: 33074657 Free PMC article.
-
T cell receptor cross-reactivity expanded by dramatic peptide-MHC adaptability.Nat Chem Biol. 2018 Oct;14(10):934-942. doi: 10.1038/s41589-018-0130-4. Epub 2018 Sep 17. Nat Chem Biol. 2018. PMID: 30224695 Free PMC article.
-
Structurally silent peptide anchor modifications allosterically modulate T cell recognition in a receptor-dependent manner.Proc Natl Acad Sci U S A. 2021 Jan 26;118(4):e2018125118. doi: 10.1073/pnas.2018125118. Proc Natl Acad Sci U S A. 2021. PMID: 33468649 Free PMC article.
-
A class-mismatched TCR bypasses MHC restriction via an unorthodox but fully functional binding geometry.Nat Commun. 2022 Nov 23;13(1):7189. doi: 10.1038/s41467-022-34896-0. Nat Commun. 2022. PMID: 36424374 Free PMC article.
-
Unravelling the intricate cooperativity of subunit gating in P2X2 ion channels.Sci Rep. 2020 Dec 10;10(1):21751. doi: 10.1038/s41598-020-78672-w. Sci Rep. 2020. PMID: 33303878 Free PMC article.
References
-
- Baker B. M., Turner R. V., Gagnon S. J., Wiley D. C., Biddison W. E. (2001). Identification of a crucial energetic footprint on the a1 Helix of Human Histocompatibility Leukocyte Antigen (HLA)-A2 that provides functional interactions for recognition by tax peptide/HLA-A2-specific T cell receptors. J. Exp. Med. 193, 551–562. 10.1084/jem.193.5.551 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

