MCPIP1 contributes to clear cell renal cell carcinomas development
- PMID: 28197812
- PMCID: PMC5511613
- DOI: 10.1007/s10456-017-9540-2
MCPIP1 contributes to clear cell renal cell carcinomas development
Abstract
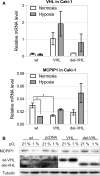
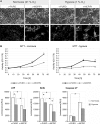
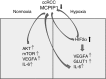
Monocyte Chemoattractant protein-induced protein 1 (MCPIP1), also known as Regnase-1, is encoded by the ZC3H12a gene, and it mediates inflammatory processes by regulating the stability of transcripts coding for proinflammatory cytokines and controlling activity of transcription factors, such as NF-κB and AP1. We found that MCPIP1 transcript and protein levels are strongly downregulated in clear cell renal cell carcinoma (ccRCC) samples, which were derived from patients surgically treated for renal cancer compared to surrounded normal tissues. Using Caki-1 cells as a model, we analyzed the role of MCPIP1 in cancer development. We showed that MCPIP1 expression depends on the proteasome activity; however, hypoxia and hypoxia inducible factor 2 alfa (HIF2α) are key factors lowering MCPIP1 expression. Furthermore, we found that MCPIP1 negatively regulates HIF1α and HIF2α levels and in the case of the last one, the mechanism is based on the regulation of the half time of transcript coding for HIF2α. Enhanced expression of MCPIP1 in Caki-1 cells results in a downregulation of transcripts encoding VEGFA, GLUT1, and IL-6. Furthermore, MCPIP1 decreases the activity of mTOR and protein kinase B (Akt) in normoxic conditions. Taken together, MCPIP1 contributes to the ccRCC development.
Keywords: Angiogenesis; Clear cell renal cell carcinoma (ccRCC); HIF1α; HIF2α; MCPIP1 (Regnase-1).
Conflict of interest statement
The authors declare that there is no conflict of interests regarding the publication of this paper.
Figures





Similar articles
-
MCPIP1 Downregulation in Clear Cell Renal Cell Carcinoma Promotes Vascularization and Metastatic Progression.Cancer Res. 2017 Sep 15;77(18):4905-4920. doi: 10.1158/0008-5472.CAN-16-3190. Epub 2017 Jul 17. Cancer Res. 2017. PMID: 28716897
-
The anti-inflammatory protein MCPIP1 inhibits the development of ccRCC by maintaining high levels of tumour suppressors.Eur J Pharmacol. 2020 Dec 5;888:173591. doi: 10.1016/j.ejphar.2020.173591. Epub 2020 Sep 21. Eur J Pharmacol. 2020. PMID: 32971087
-
MCPIP1 regulates focal adhesion kinase and Rho GTPase-dependent migration in clear cell renal cell carcinoma.Eur J Pharmacol. 2022 May 5;922:174804. doi: 10.1016/j.ejphar.2022.174804. Epub 2022 Mar 4. Eur J Pharmacol. 2022. PMID: 35257717
-
Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions.Biochim Biophys Acta. 2012 Oct;1823(10):1905-13. doi: 10.1016/j.bbamcr.2012.06.029. Epub 2012 Jul 4. Biochim Biophys Acta. 2012. PMID: 22771441 Review.
-
Activity of MCPIP1 RNase in tumor associated processes.J Exp Clin Cancer Res. 2019 Oct 21;38(1):421. doi: 10.1186/s13046-019-1430-6. J Exp Clin Cancer Res. 2019. PMID: 31639017 Free PMC article. Review.
Cited by
-
Regnase-1 downregulation promotes pancreatic cancer through myeloid-derived suppressor cell-mediated evasion of anticancer immunity.J Exp Clin Cancer Res. 2023 Oct 9;42(1):262. doi: 10.1186/s13046-023-02831-w. J Exp Clin Cancer Res. 2023. PMID: 37814340 Free PMC article.
-
Sirt3 inhibits cerebral ischemia-reperfusion injury through normalizing Wnt/β-catenin pathway and blocking mitochondrial fission.Cell Stress Chaperones. 2018 Sep;23(5):1079-1092. doi: 10.1007/s12192-018-0917-y. Epub 2018 Jun 3. Cell Stress Chaperones. 2018. PMID: 29862442 Free PMC article.
-
TAZ inhibition promotes IL-2-induced apoptosis of hepatocellular carcinoma cells by activating the JNK/F-actin/mitochondrial fission pathway.Cancer Cell Int. 2018 Aug 14;18:117. doi: 10.1186/s12935-018-0615-y. eCollection 2018. Cancer Cell Int. 2018. PMID: 30127666 Free PMC article.
-
Involvement of E3 Ligases and Deubiquitinases in the Control of HIF-α Subunit Abundance.Cells. 2019 Jun 15;8(6):598. doi: 10.3390/cells8060598. Cells. 2019. PMID: 31208103 Free PMC article. Review.
-
Network-based integrated analysis of omics data reveal novel players of TGF-β1-induced EMT in human peritoneal mesothelial cells.Sci Rep. 2019 Feb 6;9(1):1497. doi: 10.1038/s41598-018-37101-9. Sci Rep. 2019. PMID: 30728376 Free PMC article.
References
-
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Barthorpe S, Beare D, Buck G, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

