Extreme Trait Whole-Genome Sequencing Identifies PTPRO as a Novel Candidate Gene in Emphysema with Severe Airflow Obstruction
- PMID: 28199135
- PMCID: PMC5519967
- DOI: 10.1164/rccm.201606-1147OC
Extreme Trait Whole-Genome Sequencing Identifies PTPRO as a Novel Candidate Gene in Emphysema with Severe Airflow Obstruction
Abstract
Rationale: Genetic association studies in chronic obstructive pulmonary disease have primarily tested for association with common variants, the results of which explain only a portion of disease heritability. Because rare variation is also likely to contribute to susceptibility, we used whole-genome sequencing of subjects with clinically extreme phenotypes to identify genomic regions enriched for rare variation contributing to chronic obstructive pulmonary disease susceptibility.
Objectives: To identify regions of rare genetic variation contributing to emphysema with severe airflow obstruction.
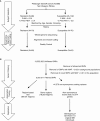
Methods: We identified heavy smokers that were resistant (n = 65) or susceptible (n = 64) to emphysema with severe airflow obstruction in the Pittsburgh Specialized Center of Clinically Oriented Research cohort. We filtered whole-genome sequencing results to include only rare variants and conducted single variant tests, region-based tests across the genome, gene-based tests, and exome-wide tests.
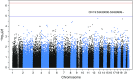
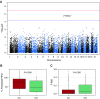
Measurements and main results: We identified several suggestive associations with emphysema with severe airflow obstruction, including a suggestive association of all rare variation in a region within the gene ZNF816 (19q13.41; P = 4.5 × 10-6), and a suggestive association of nonsynonymous coding rare variation in the gene PTPRO (P = 4.0 × 10-5). Association of rs61754411, a rare nonsynonymous variant in PTPRO, with emphysema and obstruction was demonstrated in all non-Hispanic white individuals in the Pittsburgh Specialized Center of Clinically Oriented Research cohort. We found that cells containing this variant have decreased signaling in cellular pathways necessary for survival and proliferation.
Conclusions: PTPRO is a novel candidate gene in emphysema with severe airflow obstruction, and rs61754411 is a previously unreported rare variant contributing to emphysema susceptibility. Other suggestive candidate genes, such as ZNF816, are of interest for future studies.
Keywords: chronic obstructive pulmonary disease; emphysema; genetic association studies; whole-genome sequencing.
Figures



Comment in
-
Whole-Genome Sequencing in Common Respiratory Diseases. Ready, Set, Go!Am J Respir Crit Care Med. 2017 Jul 15;196(2):121-122. doi: 10.1164/rccm.201703-0479ED. Am J Respir Crit Care Med. 2017. PMID: 28707973 Free PMC article. No abstract available.
Similar articles
-
Exome Sequencing Analysis in Severe, Early-Onset Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2016 Jun 15;193(12):1353-63. doi: 10.1164/rccm.201506-1223OC. Am J Respir Crit Care Med. 2016. PMID: 26736064 Free PMC article.
-
Integrative genomics identifies new genes associated with severe COPD and emphysema.Respir Res. 2018 Mar 22;19(1):46. doi: 10.1186/s12931-018-0744-9. Respir Res. 2018. PMID: 29566699 Free PMC article.
-
The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema.Am J Respir Crit Care Med. 2010 Mar 1;181(5):486-93. doi: 10.1164/rccm.200909-1364OC. Epub 2009 Dec 10. Am J Respir Crit Care Med. 2010. PMID: 20007924
-
Applying Functional Genomics to Chronic Obstructive Pulmonary Disease.Ann Am Thorac Soc. 2018 Dec;15(Suppl 4):S239-S242. doi: 10.1513/AnnalsATS.201808-530MG. Ann Am Thorac Soc. 2018. PMID: 30759001 Free PMC article. Review.
-
National Emphysema Treatment Trial state of the art: genetics of emphysema.Proc Am Thorac Soc. 2008 May 1;5(4):486-93. doi: 10.1513/pats.200706-078ET. Proc Am Thorac Soc. 2008. PMID: 18453360 Free PMC article. Review.
Cited by
-
Exome sequencing of extreme phenotypes in bronchopulmonary dysplasia.Eur J Pediatr. 2020 Apr;179(4):579-586. doi: 10.1007/s00431-019-03535-0. Epub 2019 Dec 17. Eur J Pediatr. 2020. PMID: 31848748
-
Accuracy and efficiency of germline variant calling pipelines for human genome data.Sci Rep. 2020 Nov 19;10(1):20222. doi: 10.1038/s41598-020-77218-4. Sci Rep. 2020. PMID: 33214604 Free PMC article.
-
Identifying Chronic Obstructive Pulmonary Disease Genes: Shining the Light on Dark DNA.Am J Respir Cell Mol Biol. 2019 Apr;60(4):373-374. doi: 10.1165/rcmb.2018-0349ED. Am J Respir Cell Mol Biol. 2019. PMID: 30376354 Free PMC article. No abstract available.
-
Assessing the contribution of rare genetic variants to phenotypes of chronic obstructive pulmonary disease using whole-genome sequence data.Hum Mol Genet. 2022 Nov 10;31(22):3873-3885. doi: 10.1093/hmg/ddac117. Hum Mol Genet. 2022. PMID: 35766891 Free PMC article.
-
Prognostic signature of lung adenocarcinoma based on stem cell-related genes.Sci Rep. 2021 Jan 18;11(1):1687. doi: 10.1038/s41598-020-80453-4. Sci Rep. 2021. PMID: 33462260 Free PMC article.
References
-
- Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, et al. Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. - PubMed
-
- National Heart, Lung, and Blood Institute. Morbidity and mortality: 2012 chart book on cardiovascular, lung and blood diseases. Bethesda, MD: National Institutes of Health; 2012 [updated 2013 May; accessed 2016 May 21]. Available from: https://www.nhlbi.nih.gov/research/reports/2012-mortality-chart-book.
-
- Shapiro SD, Ingenito EP. The pathogenesis of chronic obstructive pulmonary disease: advances in the past 100 years. Am J Respir Cell Mol Biol. 2005;32:367–372. - PubMed
-
- Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. ICGN Investigators; ECLIPSE Investigators; COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–957. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

